Content from Introduction
Last updated on 2024-08-13 | Edit this page
Estimated time: 20 minutes
Overview
Questions
- What should I expect to learn from the HPC User module?
Objectives
- Set the basis for learning about High-Performance Computing in Science.
Overview
In doing computational science it is very common to start a project by writing code on a personal computer. Often as the project proceeds we find that we need more computer resources to complete the science project. This may come in the form of needing more processing power to complete the research in a reasonable time. It may also mean needing more memory to be able to run larger calculations. Or it may just mean needing to do a very large number of smaller jobs that would overwhelm a single computer system.
In these cases where we need to seek out more computational resources, we also need to start understanding the performance aspects of our code. More power is not always the answer, sometimes writing more efficient code can get the job done equally as well.
This HPC User lesson is aimed at scientists who need to use computers to do calculations, and not at computer scientists or computer engineers who need to be experts at programming in a High-Performance Computing environment. This lesson will be aimed at giving an overview of performance concepts to provide a general understanding of how to operate in an HPC environment.
Organization
The first few chapters concentrate on discussing performance issues at the conceptual level with practical examples. These examples are currently given in Python but it is intended to eventually have the user and instructor choose the language that the examples display in to make it more appropriate to teach this to groups primarily interested in R, C/C++, Fortran, or Matlab too. As the lesson proceeds these same concepts will be used in different ways and with examples in different computer languages to help drill them in.
The middle third of the lesson is a language survey. Even though most scientists may work primarily in a single language, it is important to understand the strengths and weaknesses of alternative languages as well as their own favorite.
The last sections provide overviews of some more advanced topics like working with GPUs to accelerate scientific codes. It may be that some of this will be skipped by your instructor due to time limitations but it is good to have these available for reference purposes.
There are hands-on exercises throughout the lesson where you will be asked to apply some of what you have learned. There are also optional homework assignments available for those who want to challenge themselves outside of the workshop.
Most sections also have website links at the end which provide a means to seek out more information.
- The HPC User lesson will help to understand basic concepts affecting performance in programming.
Content from Profiling Code for Performance
Last updated on 2025-09-17 | Edit this page
Estimated time: 20 minutes
Overview
Questions
- How to measure performance?
- How to measure memory usage?
Objectives
- Learn the different methods available to measure time and memory usage externally and from within a program.
- Understand what parts of a program are important to time.
- Learn how to do a scaling study for multi-core and multi-node jobs.
When we talk about the performance of a program, we are always interested in how much time it takes to run, but in some cases we also need to know how much memory the program uses if we are pushing the limits of our computer. Even more than that, we often need to know how much time each part of the code takes so that we know where to concentrate our efforts as we try to improve the overall performance of the program. So we need to be able to time the entire run, and also internally each part of the code. For parallel codes, we also need to know how efficiently they scale as we increase the number of cores or compute nodes in order to determine what resources to request.
Timing a Program Externally
Linux has a time function that can proceed any command, so this can be used to time the entire job externally even if we don’t have access to the source code. This can be used to time a short test run in order to estimate the run time needed for the complete job. When we get to talking about parallel computing, or using multiple compute cores to run a single job, the time function will prove useful for getting the execution time for a job as it depends on the number of cores. For example, it is common to test the same job on 1, 4, 8, 16, and 32 cores to see how it scales with more cores to determine what number of cores is most efficient to use.
Let’s start with a few simple examples of using the time function at the command prompt. Try timing the pwd command which prints the current working directory.
OUTPUT
/Users/daveturner
real 0m0.000s
user 0m0.000s
sys 0m0.000sThe first thing you see is the output of the pwd command, which in this case is the directory /Users/daveturner on my Mac. Then the time function prints its output, always as real time, which is what we want, then user and system time which we can ignore. This shows that the pwd command is faster than the clock can measure. This is important to note that the time command isn’t accurate to less than 1 millisecond, so in general we should always make sure we are measuring execution times that are greater than a second in general.
Below is another example where this time we are timing the ls function which will list the files in the current directory.
OUTPUT
file1 file2 file3
real 0m0.110s
user 0m0.006s
sys 0m0.027sWhen I do this on my Mac, I get a real time of just 0.006 seconds because it’s really fast to access the local hard disk. The test above is from a very large cluster computer that has a parallel file server with 1 Petabyte of storage (1 Petabyte is 1000 Terabytes, and each Terabyte is 1000 Gigabytes). The performance for accessing large files on a parallel file server is very good, but it does take longer to do small tasks like get the contents of the current directory. How does this compare to the system you are on? On large HPC (High-Performance Computing) systems like this cluster, the speed also depends on what file system you are testing. You can usually access the local disk at /tmp, but your home directory may be on another file system, and often there is fast scratch space that is very high performance but only used for programs when they are running.
Below we are going to use the sleep command to time an interval of 5 seconds just to simulate what we might see in a real job. In this case, even though the sleep function was supposed to go 5 seconds, there was some overhead or inaccuracy in the timing routine or the length of the sleep. This is one thing that you always need to be aware of when measuring performance. If you are measuring short things, you may want to measure multiple times and take an average.
OUTPUT
real 0m5.052s
user 0m0.001s
sys 0m0.003sIn addition to worrying about the clock accuracy, you also need to worry about interference from other jobs that may be running on the same compute node you are on. The best way to time a real job is to test it out on a completely isolated computer. If you are on an HPC system with a batch queue, you can always request an entire compute node and then just use part of the node you requested, even a single compute core. If this is not possible, then try to get the job as isolated as you can. Submitting a single-core request to a job queue is one example of this, where you at least are sure that your job is the only one on the cores that you requested. Your job will still be sharing the memory bus and L3 cache with other jobs. Jobs usually don’t effect each other much from this, but they can. If your job is using the network to communicate with other compute nodes, that might also be shared with other jobs running on the same node. The single largest factor to be aware of is that other jobs using the same file server as you are can definitely affect the performance of your job if your code is doing lots of IO (Input and Output). On HPC systems, this can be true even if the other jobs are not on the same compute node as your job. If you want to isolate your job from others in this case, you will need to do your IO to the local disk (usually /tmp).
Timing Internal Parts of a Program
Every computer language has multiple clock functions that can be used to time sections of the code. The syntax is different in each language, but they all work about the same, and there is always a very high precision function that is accurate down to somewhere in the nanosecond range, though I typically don’t trust these for measuring less than a microsecond interval. Each of these clock functions returns the current time, so to measure the time in an interval you need to store the start time, do some calculations or IO, then get the end time and subtract the two to get the time used for that part of the code.
In Python since version 3.3, the best timer is in the time package. To use it, you must start by importing the package. In C/C++ the clock_gettime() function returns the current time accurate to less than a microsecond. Accuracy of other timers like SYSTEM CLOCK() in Fortran 90 vary with the system and compiler. Below are examples of using the clock functions to measure the time it takes to do a loop, then measure the time it takes to dump an array out to a file.
Try running the timing_example code yourself for the language you are working with. These are codes you should have downloaded and unzipped in your HPC system, and should be in the code directory. Timing will be dependent on the language, but the values I see are in the millisecond range. Since both of these are above the nanosecond range, we can be confident that the timing routine is accurately measuring each.
Let’s see what we can learn by playing around with it some more. When I run the python version preceded by the Linux time function, I see a real time significantly larger than the loop time and output time combined. The initialization time is not measured but shouldn’t be more than the loop time. What all this means is that there is some startup time for getting the python program running and importing the time package, but we may also be seeing the lack of accuracy of the external time function when it comes to measuring things down in the millisecond range. We do not see the same time discrepancy when running the C version.
Now lets change the program itself. Sometimes we need to time a part of something that is in a larger loop, so we need to sum the times together. Try changing the timing so that it is inside the summation loop instead of outside it to see what happens. You can do this by uncommenting the timing and printing functions in the code file.
In my computer, the t_sum time is only a bit larger than the t_loop time from before, but remember that this doesn’t count the loop overhead. If we look at the t_loop time instead, in my computer it is more than double what it was before. When the clock routine is measuring very small intervals each time, it can be intrusive in that it distorts the measurement by increasing the run time of the entire code. It isn’t surprising that this is intrusive since we are measuring the time it takes to retrieve a single array element and do one addition. The code is doing a subtraction and addition itself to calculate the time interval, so it is probably more surprising that doing the timing in this way is not more intrusive.
What to Time
The goal is to fully profile your code so that you understand where all the time is being spent. This means timing each computational section where time is being spent, usually the loops for example. While simple print statements may not be important contributors to the overall run time of a code, any large input or output from files may be. When we start talking about parallel programs that use multiple cores or even multiple compute nodes it will become important to measure the time taken in communicating data to other cores and other nodes. Once you have a complete profile of where time is being spent, then you can understand where to start in trying to optimize your program to make it run faster.
Measuring Parallel Job Scaling
When we get to talking about multi-processor jobs it will be very important to understand how efficiently a job scales as we use more processing cores. For this we will do what is called a scaling study where we measure the performance of a typical job using different number of processing cores. We may for example run on 1 core, then 4, 8, 16, and 32 cores to see how efficiently the job scales as we apply more processing power. This is done so that we can understand how many cores we can efficiently apply to that job. If we get a 7 times speedup on 8 cores compared to 1, that less than ideal but still very good. If we then see a 9 times speedup using 16 cores, then we’d probably want to stick with using just 8 cores on that particular job. Do keep in mind that scaling is very dependent on the problem size. Larger problems will typically scale better to more cores, while smaller problems may be limited to only a few cores. The goal is to determine how many cores we can use with reasonable efficiency. The same kind of scaling study can also be used for multi-node jobs, where we would test the performance on 1 node, 2, 4, and 8 nodes for example.
Problem size is one factor that can affect the scaling efficiency. For multi-node jobs, choosing the fastest networking options, and ensuring that all compute nodes are on the same network switch can both increase the scaling efficiency.
Tracking Memory Usage
When we think about high performance, we mostly think about running jobs faster. For some programs, the memory usage may be the factor limiting what types of science we can do. At the very least, we need to know what memory to request when submitting jobs to a batch scheduler.
For simple jobs, like the matrix multiplication code in the next section, we can calculate the exact memory requirements. If we are going to multiply two matrices of size NxN and put the results in a third, then each matrix takes NxN x 8 Bytes if the elements are 64-bit floats, so the total memory required would be 3 x NxN * 8 Bytes.
For more complicated programs, often the best approach is to do a short test run to measure the memory use before submitting the full job. This is especially true if you are submitting lots of similar jobs. If your job goes over the requested memory, it is most often killed, so you want to over estimate the request somewhat, but if you request too much it can take a lot longer to get your job scheduled and result in inefficient use of the resources as you will be locking up memory that you are not using.
Measuring the memory usage is more difficult than it should be on most systems, and it depends on what tools you have available. If you are on an HPC system with a batch scheduler like Slurm, you can use the sstat command to find the current memory usage for a running job and seff to find the maximum memory used for a completed job, using the job ID number in both cases to choose the job you are interested in.
OUTPUT
Job ID: 5072064
Cluster: beocat
User/Group: daveturner/daveturner_users
State: FAILED (exit code 16)
Cores: 1
CPU Utilized: 00:01:07
CPU Efficiency: 97.10% of 00:01:09 core-walltime
Job Wall-clock time: 00:01:09
Memory Utilized: 2.26 GB
Memory Efficiency: 11.31% of 20.00 GBThe matrix multiplication job above used 10,000x10,000 matrices and took 1 minute 9 seconds. We see that in this case it used 2.26 GB of memory. One thing to keep in mind is that 1 GB can be calculated in different ways. In this case, 1 kB is 1024 Bytes, 1 MB is 1024 x 1024 Bytes, and 1 GB is 1024 x 1024 x 1024 Bytes. So 3 matrices are 3 x NxN x 8 / (1024x1024x1024) GB = 2.235 GB, rounded up to 2.26 GB. In general, if you estimate 1 GB as 10^9 Bytes that works fine.
OUTPUT
MaxRSS
----------
2289.50MFor a running job you can use the sstat command with the job ID number. The sstat command will dump a lot of information out, so using the –format=MaxRSS parameter provides just the real memory that we want. In this case, it is again 2.29 GB.
Different HPC systems may have batch queue systems other than Slurm, but all will have similar methods of getting the memory usage for running and completed jobs. If you are not running a job through a batch system, you can use the htop command to find your process and look at the Res or resident memory. This works best for single-core jobs as multi-core jobs may show memory usage for each thread.
Ganglia is another option providing a web-based interface to look at memory usage over time. If you have access to Ganglia on your HPC system, it provides a node view only rather than a job view, so it is only really useful if your job is the only one running on a given compute node. However, being able to see the memory use over time can be very helpful.
We will practice these approaches more in the upcoming modules.
- The time function can always be used externally to measure performance but has limited accuracy of around 1 millisecond.
- Internally there are precise clock routines that can be used to measure the performance of each part of a code. These are different for each programming language, but the use is always the same.
- Scaling studies can help determine how many cores or nodes we can efficiently use for a parallel job of a given problem size.
- Measuring memory use can be done from outside a program, but you may also be able to calculate total memory for simple programs.
Content from Performance Concepts
Last updated on 2025-10-07 | Edit this page
Estimated time: 40 minutes
Overview
Questions
- What does performance mean in programming?
- How do I take advantage of optimized libraries?
Objectives
- Understand that what actually goes on in the computer may be much more complex than what the user tells the computer to do.
- Have a basic understanding of some of the performance issues to be aware of.
- Learn that you don’t have to be an expert programmer to take advantage of advanced performance techniques, you just need to be aware of how to use libraries optimized by experts.
Conceptual View
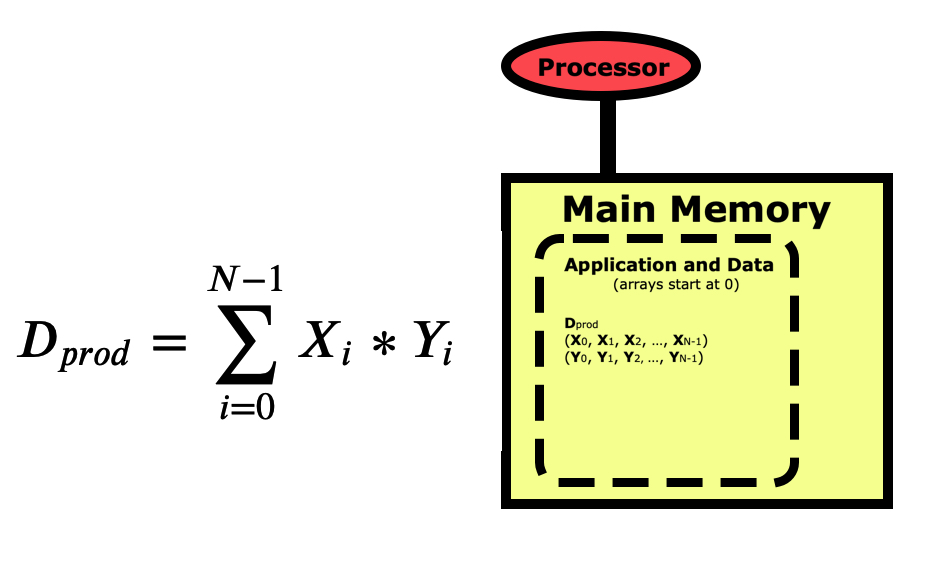
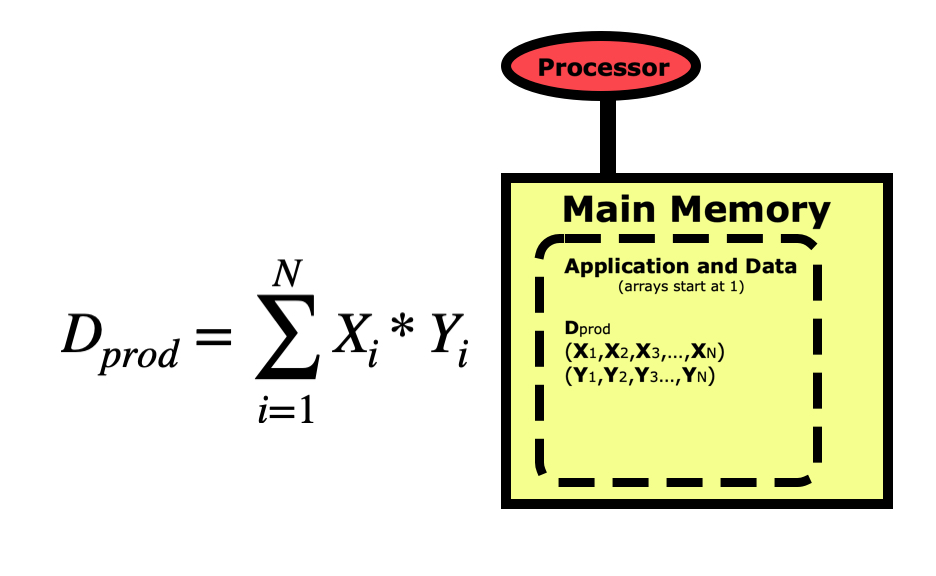
When we write a program for a computer, we view the operation from a more conceptual level. The picture below is for a simple dot product between two vectors X and Y, where the dot product Dprod is the sum of the elements of each vector multiplied together. Notice first that the indexing for arrays in Python starts at 0 and goes to N-1. This varies between languages, with C/C++ also starting at 0, while R, Matlab, and Fortran start arrays at 1 and go to N.
When we think of a computer running a program to do this calculation, we view it as starting with the variable Dprod being pulled from main memory into the processor where this sum is zeroed out. We then pull the first element of X and the first element of Y into the registers and multiply them together then summing them into Dprod. We loop through all elements of the vectors, each time pulling the X and Y element into memory, multiplying them together and summing them into Dprod. Then at the end, we push Dprod back down into main memory where the program prints the result out to the screen.
This conceptual view of what the computer is doing is all we really need to be aware of when we are starting to writing programs. But when those programs start taking too long to run on a personal computer then we need to understand what the computer is doing in more depth so we can make sure that the code is running optimally. Computers are internally quite complex, so fully understanding how code can be written in different ways to streamline the processing can be very challenging. Fortunately, not everyone needs to be able to write optimal low-level code. For most of us, we just need to understand what programming techniques in each language may cause performance problems, and understand how to take advantage of optimization libraries that experts have written for us.
The Computer’s View
Let’s walk through the same dot product example again, but this time looking at it from the point of view of the computer rather than our conceptual view. The first thing to understand is that when you pull a variable up from main memory into the registers in a processor, what goes on behind the scene is very complicated, but fortunately for us also totally automated. Variables don’t get pulled up individually but as part of a 64-byte cache line. The variables in this cache line get promoted through several layers of increasingly fast memory known as cache layers, with a copy of the entire cache line being left behind in each layer. In this way, the most frequently used data will be more likely to be in one of the cache layers where it can be accessed more quickly. In the case of our dot product, that means loading the first element of X and its 64-byte cache line may take 10-33 ns while the next 7 only take 0.3-1 ns since those elements are now in L1 cache too.
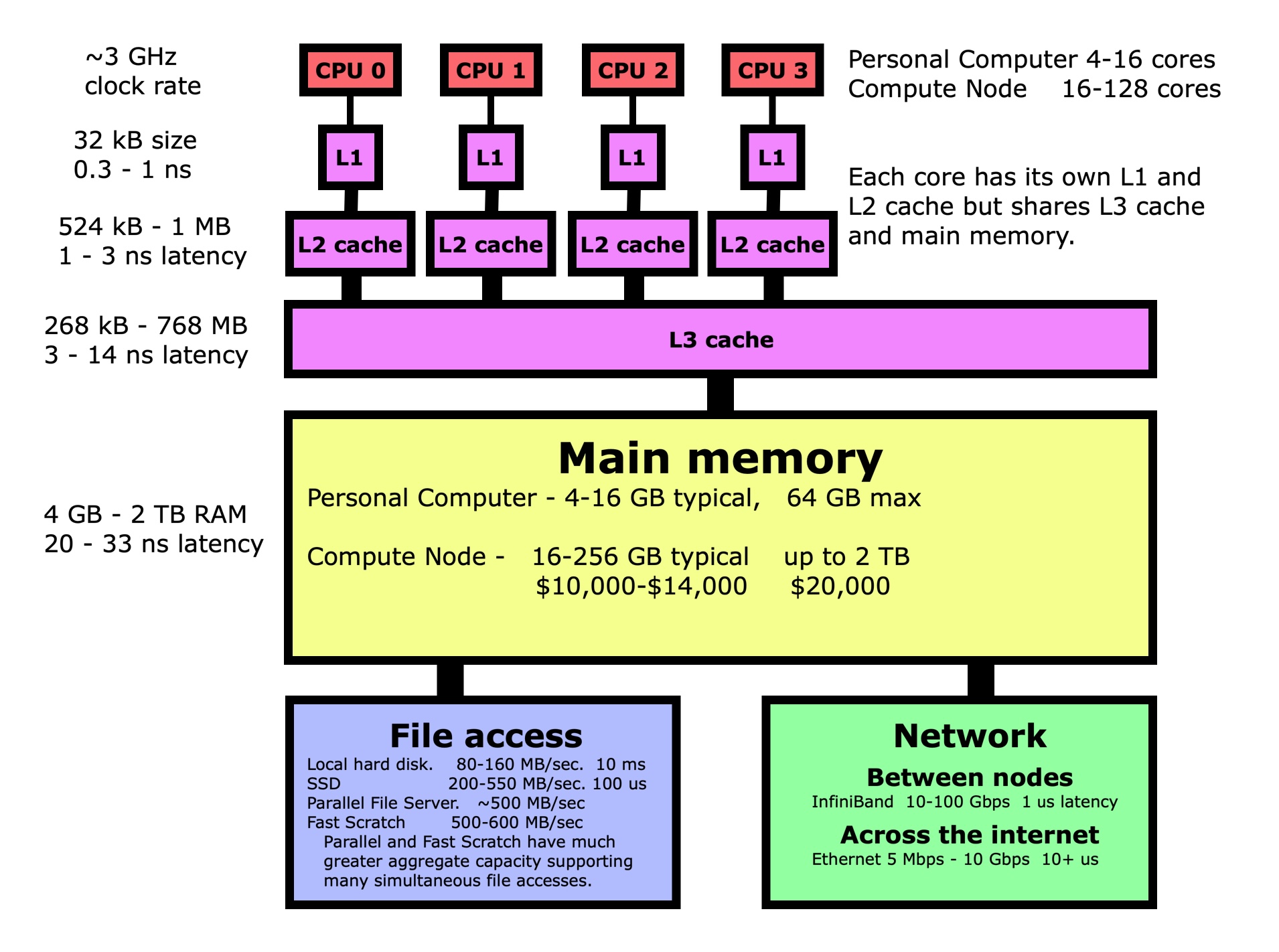
How does this knowledge help us? Performance is more about getting data to the processor since most operations are very fast once the data is in the processor registers. If the vector is instead not stored in contiguous memory, with each variable in the next memory location, then the subsequent 7 memory loads of X and Y will take 20-33 ns each instead of 0.3-1 ns each. This means that we need to ensure that variables we are using are in contiguous memory whenever possible. There will be an exercise at the end of this section where you will measure the difference in execution time between these two cases.
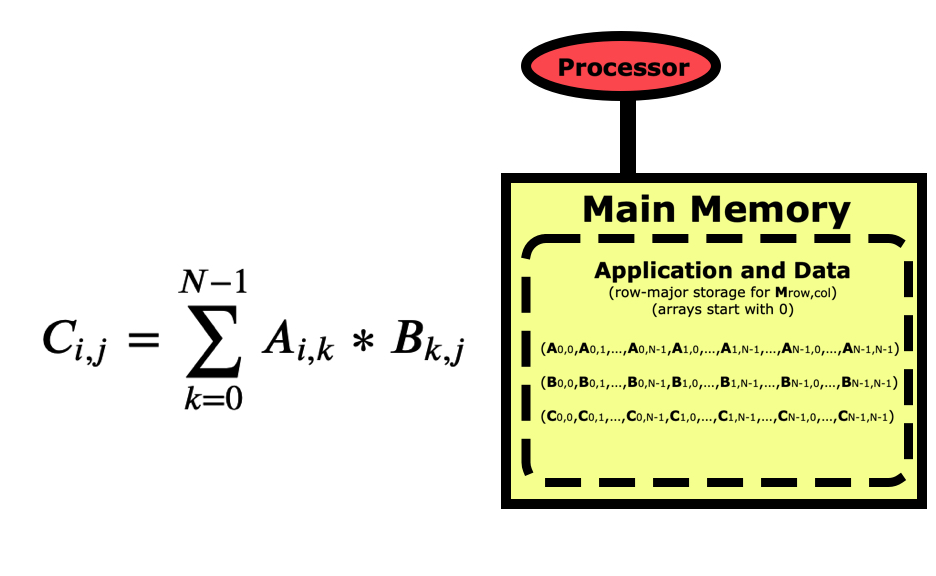
Now let’s look at a simple matrix multiplication algorithm which is fairly simple to program, but may have very different performance depending on how you write the code.
Once the matrices are initialized, the code to multiply them together is fairly simple.
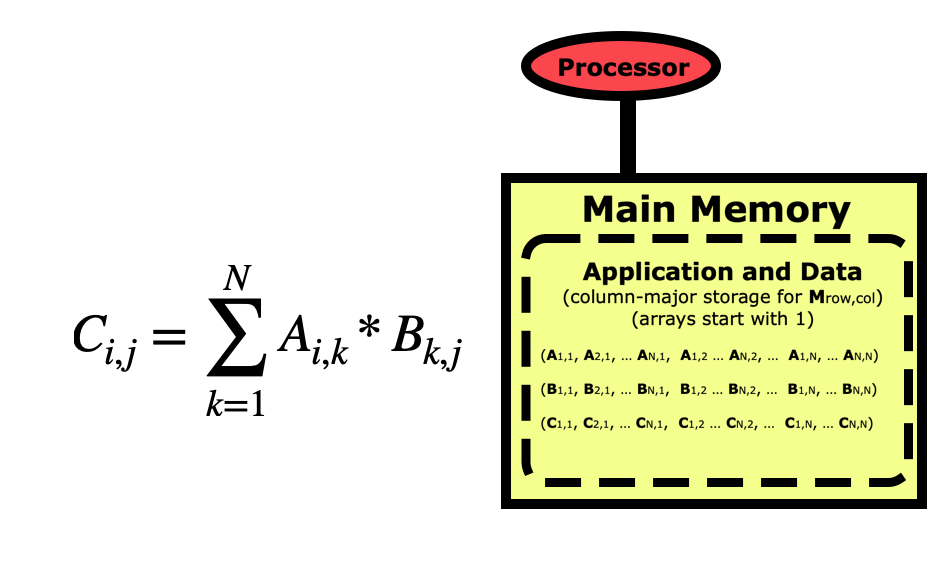
But if we are concerned about performance, we need to take a better look at this code. Python is a row-major language like C/C++, which means matrices like this are stored by rows first as in the picture above. If we look at this code from a cache-line point of view, then it looks optimal for the B matrix since when we load an element the others in the cache line will get used in the next loop iterations, but the A matrix is the opposite where elements being used next are farther apart.
But it turns out this isn’t really the way to look at performance in this case. For each element of C that we calculate, we will need N elements of A and N elements of B. So every element of A and every element of B will get re-used N times during this calculation. What is important is that when we pull each element into the L1 cache where it is the fastest to access, we want to re-use it as much as possible rather than having to pull it repeatedly from the lower caches or main memory.
An optimal approach has been developed in the past that pulls blocks of each matrix into L1 cache. This block optimization results in a much more complicated code, but also a much higher performing one. The good news is that for users like us, we don’t have to ever program it ourselves, we just need to know that it is in optimized libraries that we can call at any time. Below are some examples of how to access the low-level optimized libraries in the various languages.
So while writing a matrix multiplication code from scratch is not difficult, using the optimized function accessible from each language is easier and guarantees the best performance. To find out how much difference there is in performance, you will need to try for yourself by measuring the execution time for a few different matrix sizes in the exercise below.
Measuring Cache Line Effects
Time Different Matrix Multiplication Methods
Advanced Exercise - Transpose B to cache-line optimize it
As k is incremented in the innermost loop, elements of A are being brought into cache efficiently since they are stored in contiguous memory, as we learned from the dot product example. Unfortunately, elements of B are not since they will be sparse, separated by N-1 elements each time. While in most languages we already know that we can simply use the optimized library routine speedup the code, if this wasn’t available one thing we’d consider is to transpose the B matrix so that it is stored in column-major format before doing the matrix multiplication. If you’re up for a challenge, try programming this up to see if it improves the performance compared to the original code.
There are a great many levels of optimization that can be done to the matrix multiplication algorithm. Transposing B should improve the performance some. For a more complete overview of what is done in the np.matmult() algorithm and others like it, follow the link: https://en.algorithmica.org/hpc/algorithms/matmul/
Summary
When it starts taking longer to run a given program, we need to start looking beyond just whether it gives the correct answer and begin considering performance. This means we need to go beyond the conceptual view of how the program runs to look at how it makes use of the underlying hardware architecture. While this can get very complicated very quickly, most users just need to be aware of where performance bottlenecks can occur in order to avoid them. Quite often, writing optimal code just means taking advantage of highly optimized libraries that experts have written and tuned. So most of us just need to know when and where to look for these optimized routines in order to write highly optimized code.
- A computer’s view of code is more complex than a user’s view.
- It’s important to understand a little about what goes on when code actually runs, but you don’t need to be able to program at that level.
- Whenever possible, use code written and optimized by experts instead of writing your own version.
Content from Parallel Computing Concepts
Last updated on 2025-09-12 | Edit this page
Estimated time: 40 minutes
Overview
Questions
- Now that we can profile programs to find where the time is being spent, how do we speed the code up?
- What is parallel computing, and what are the underlying concepts that make it work?
Objectives
- Learn different approaches to speeding up code.
In the last chapter we learned how to profile a program to understand where the time is being spent. In this section, we will talk more about how to improve the performance of different parts of a program. This may involve steering clear of inefficient methods in some computer languages, minimizing the operation count, using optimized libraries, and applying multiple cores or even multiple compute nodes to speed up each computational part. Computations can sometimes be sped up enormously using accelerators like GPUs (Graphic Processing Units) as well. Reducing or eliminating IO can sometimes help, but ensuring that the IO is done without costly locks, and to a file server that is not overly used, can often help performance greatly as well.
Optimizing Scalar Code
Programming in languages like C/C++ and Fortran produces fairly efficient code in general because these are compiled languages. In other words, you need to compile the code before executing it, and the compilers do very intricate optimizations to ensure the resulting executable is highly efficient. Interpretive languages like Python, R, and Matlab only do some compilation on the fly. They are therefore much less optimized, but more convenient to run. We have already learned the importance of using optimized library routines whenever possible, but this is especially true for the interpretive languages. Some languages also have certain methods that are convenient, but very inefficient, and what to avoid and how to get around them will be discussed in later chapters.
One thing that can help in understanding performance is to know how much it costs to perform different common math functions. We can express the speed of a code in GFlops, or Giga (Billion) Floating-point operations per second. A floating-point operation involves two operands that are typically 64-bit floats. When counting the Flops, we ignore integer arithmetic and comparisons since those are very fast in relation to the floating-point operations. Below is a table of the Flop cost for each operation. This can be thought of as for example how many operations does it take to do a cosine function, since the cosine is done using a math library.
| Function / Operation | Flops count |
|---|---|
| * + - | 1 |
| / | 4 |
| square root | 4 |
| sin()/cos()/tan() | 6 |
| exponent() | 14 |
One example of how this knowledge can help is if you have a large loop where you are dividing by 4.0. Instead, you reduce the operation count by 3 if you multiply by 0.25. This is not needed in compiled languages like C/C++ or Fortran since the compiler does this optimization for you, but it can help in the interpretive languages. The example below reduces the floating-point operation count by removing the redundant calculation of theta and replacing the expensive calculations of the cosine and sine by using an iterative method using trigonometric identities.
This is however a very over-simplified picture since it involves just analyzing one factor. In practice, most processors can overlap calculations for better speed like in the AVX vector instruction sets of the Intel and AMD x86 architectures.
Flops count
Count the floating-point operations (Flops) in the unoptimized and optimized versions of the code above and calculate the expected speedup rate.
The unoptimized code uses N_atoms * ( 1 + 21 * NQ ) Flops. The optimized version uses N_atoms * ( 13 + 6 * NQ ) Flops. For large NQ the speedup would be around 21/6 or 3.5 times.
The compiled languages C/C++/Fortran would optimize some of this loop automatically by pulling the 2.0 * PI * dx / NQ out of the NQ loop and just multiplying this by iq each time.
Parallelizing Code
It is always good to optimize a scalar code first, but if you still need your code to run faster then one option is to use more processing power. This means using more then one computational core with each core working on a different part of the data. The goal is to get a speedup of N times if you are using N cores, though often parallel programs will fall short of this ideal.
There are two main types of parallel programming. The first is multi-core or shared-memory programming. This is the easiest approach, with a single program running but for computationally demanding sections like loops multiple threads are used with typically one thread on each computational core. In this way, the scalar part of the code does not need to be altered and we only need to put parallelizing commands before each loop to tell the computer how to divide the problem up. It is called shared-memory because all threads operate on the data that is shared in main memory. This is the easiest approach, is often very efficient, but has the limitation that it needs to work only on a single compute node. Multi-threading in C/C++ and Fortran use the OpenMP package, Python uses a simplified version of this called pymp, and R allows for this through the mclapply() function.
If you need even more computational power, or need access to more memory than is on a single node, then you need to use multi-node computing. This is also referred to as distributed-memory computing since each thread in this case is its own separate but identical program, again just operating on a different part of the data. Distributed-memory programs can be run on a single compute node when needed, but are designed to run on very large numbers of nodes at the same time. Some programs have been run on millions of compute nodes, or some of the largest HPC systems in the world which may cost more than $100 million. C/C++ and Fortran codes use MPI, the Message-Passing Interface, launching all the copies of the program to the different nodes using the mpirun command, then each node shakes hands with the others with the MPI_Init() function. Each thread or task will operate on a different part of the data, and when data needs to be exchanged the programmer can use MPI commands like MPI_Send() and MPI_Recv() to pass blocks of data to other threads. This is a very powerful way to program, but it is definitely much more complicated too. Python has the mpi4py package which is a stripped down version of MPI, but unfortunately you cannot do multi-node computing with R.
You will not be taught how to program in these parallel languages in this course, but you will be shown how to recognize each type of parallel approach and how to work with each efficiently.
Parallel Computing Concepts
The syntax for doing parallel processing is different for multi-threaded and multi-node programming, and also can vary for each language, but handling multiple threads at the same time always involves some of the same basic underlying concepts. Understanding the basic concepts underlying these methods will help us to understand the functions at the language level themselves.
Locks in Programs and File Systems
A multi-threaded program uses shared-memory where many threads may want to access the same data at the same time. If they are all reading the data, this is not a problem. However, if even one thread wants to change the data by writing to it while other threads may be reading it, this can lead to uncertain results depending on which thread does its read or write first. This uncertainty must therefore always be avoided, and often it is handled by locking memory when a write occurs so that only that one thread has access at that time.
The same thing can happen in parallel file servers where there are multiple paths being exploited to the same data file in order to provide better performance. If multiple threads, or even multiple programs, are reading the same file or different files in the same directory then everything is fine. However, if one of those is writing to a file then a parallel file server will lock the entire directory down to prevent other threads from reading until the write is completed. This is something that every user needs to be aware of. It only applies to parallel file servers, so local disk (/tmp) has no problems with this since there is only one controller for the disk, while a parallel file server has many controlling nodes. Some scratch space also can handle multiple writes to the same directory without locking. Since this can have severe impacts on performance, it is always good to ask your system administrator if you don’t know. Ways around this include switching to a different file system like /tmp while running the job, or putting the files in different directories.
Barriers
Since distributed-memory programs involve multiple copies of the same code, we commonly need to ensure that all are at the same point in the code. MPI uses barriers for this, an MPI_Barrier() function to be exact. When this is encountered, each task will stop and wait until all tasks reach that point in the code, they will communicate this to each other, then continue on. A common example of why you would need this would be in debugging an MPI code where you want to identify where the code may be failing. If one task gets ahead of the other and errors out, it may be that the root task will be at a different place in the code and report that line where the job failed.
Forks
All multi-threaded packages use some sort of a fork function. When a loop is encountered and the root thread needs to spin up multiple threads to do the work, it does so by doing a fork() which duplicates the variables in the root thread. This is done virtually which may be a bit confusing. If every piece of data was copied it would increase the memory usage enormously for runs on large numbers of cores, so only the pointers to the arrays are copied. If the data is only being read then all threads can read from the original array. If any thread writes to part of an array, then a unique copy of that part of the array is made for that thread only. So the fork process manages the memory usage behind the scenes to minimize the redundant storage of data, but ensures that there is no need for a memory lock when a thread writes to that data by making a copy instead.
Dependencies in Loops
All those mechanisms discussed above may be used in implementing a parallel computing package. As a user, what we really need to know is when can a section of a program be parallelized. If you look at the loops where the most computational time is being spent, what you need to determine is whether each pass through the loop is independent of the others, or whether each pass is dependent on the results of the previous iteration. If each pass through a loop can be done independently of the others, then we can do them at the same time. This is a simple statement, but it does sometimes take more thinking to understand if there are any dependencies involved. If you have any doubt, try writing down all the variables that are needed as input for each iteration of the loop, then see if any of those change throughout the loop.
If you have a program with nested loops, you may need to analyze each loop level to see if it is parallelizable. Parallelizing the outer loop means that there will be more computations for each thread or task, which is referred to as being more coarse grained. This can lead to much higher efficiencies, but it is not always possible. Often it is the inner loop that is easiest to parallelize, and this is most often the case with multi-threaded parallelism.
Using Accelerators like GPUs
Some programs can be sped up by using a GPU as a computational accelerator. A 32-bit GPU is the same as you would buy for a high-end gaming computer and can cost $1000-$1500. These are ideal for accelerating 32-bit codes like classical molecular dynamics simulations, and have custom hardware that is great for training AI (Artificial Intelligence) neural networks or machine learning models. The more expensive 64-bit GPUs are never intended for graphics at all. They are custom designed as accelerators even though they are still called GPUs. These currently cost around $11,000 for an NVIDIA A100 and around twice that for a newer H100.
Writing a program to run on a GPU is very difficult. For NVIDIA GPUs, you use a programming language called CUDA. There are many fewer codes optimized for AMD GPUs at this point. They are programmed with Hip which can be compiled to run on either AMD or NVIDIA GPUs. There are also projects in development to convert native CUDA codes into executables optimized for AMD GPUs.
Running a job with a GPU accelerator is not that difficult. If your application can make use of one or more GPUs, there will be directions on how to specify the number of GPUs. If you are on an HPC system, you can request the number and type of GPUs you want for each job.
Optimizing Input and Output
The first thing to understand about IO (Input and Output) is that it can make a big difference as to what type of a file system you are reading from or writing to. Local disk (usually /tmp) is temporary storage and has size restrictions, and it isn’t as fast as a parallel file server that stripes data across many disks at the same time, but it is still sometimes the best to use if others are heavily using the main file server and slowing it down. As good as parallel file severs are, they also commonly need to lock a directory if more than one file is being written at the same time. Any locking can slow the performance of a code down immensely and should be avoided if at all possible. Many HPC systems may have fast scratch space which is temporary storage often purged every week or month but very large in size. This is designed for use when you are running your job, and may also not suffer from the same locking issues as on some parallel file servers.
On our HPC system at Kansas State University, our fast scratch is about ten times as fast as the parallel file server system that our home directories are on. So you would think that all you have to do is use fast scratch all the time to make your IO ten times faster. It actually is the case if you are streaming data in, by which we mean reading in data in large chunks that do not need to be converted. Files with large strings like genetic information fall into this category since the strings can be hundreds or thousands of characters long and the representation in the file is the same as in the program. Reading in arrays of floats or integers from binary files also can go as fast as the file server allows since the elements are stored in binary in both the file and the program.
The problem comes when we want to store numbers for example in a text file so we can see them ourselves. When we write them or read them, the process goes slow since we have to convert each number from its binary representation into a text string before reading or writing. With IO, it is that conversion process which is slow, so it doesn’t matter how fast the underlying file server is in these cases. So if you want to speed up IO, think about streaming data in binary form if possible, and if so then choose the fastest file server available.
Scaling Study of the Distributed-Memory Dot Product Code
Measure the entire run-time for the dot_product_message_passing code for the language you are working with for 1, 4, 8, and 16 cores if you are on an HPC system with at least 2 compute nodes. You can try different combinations of nodes and cores for each if you would like to see the effects of the network (for the 4 cores, try 2 nodes 2 cores vs 4 nodes 1 core).
In this code we initialize the vectors locally so there is no communication involved. The only communication is the global sum at the end, so we expect the scaling to be close to ideal. In many practical MPI codes, we would need to read data into one of the ranks, divide the data up and send it out to each other node. Real MPI applications also usually require communication mixed in with the computational part in order to get data where it needs to be. All this communication can slow down the job, and this usually gets worse as you spread a job over more cores, and especially over more nodes.
- What techniques can be used to speed up scalar code?
- How to improve input and output?
- Learn about the difference between multi-core and multi-node programs.
- Understand the fundamentals of locks, barriers, and forks.
- Practice doing a scaling study.
Content from Multi-Threaded Programs
Last updated on 2025-09-17 | Edit this page
Estimated time: 30 minutes
Overview
Questions
- What is the multi-threaded shared-memory programming model?
Objectives
- Learn about multi-threaded computing and how to use it.
- Understand the strengths and limitations of this approach.
Most computer languages have the ability to do multi-threaded computing. C/C++ and Fortran use the OpenMP package which is by far the most extensive and well developed. It uses pragma statements to control the parallelization of loops so that multiple compute cores work at the same time on different parts of the data. OpenMP is not only extremely efficient, but it also provides very advanced features offering greater control on how the parallelization is to be done, all without encumbering the programmer too much. The pymp package for Python is a stripped down version of OpenMP supporting just the basic functionality. It is an actively developed project and is very efficient and relatively easy to use as well. R takes a very different approach in doing multi-threading using the mclapply() function which is a multi-core replacement for the lapply() function. This operates similarly to OpenMP and pymp but uses a very different syntax. It is also not nearly as efficient and requires some non-default choices to make it more perform better. Matlab also uses a different syntax in its Parallel Computing Toolbox where it uses a parfor command to do a parallel for loop.
All of these methods behave basically the same by forking, or splitting off, extra compute threads when called. Each thread gets its own virtual memory space, meaning most large arrays are not copied during the initialization stage. If any thread writes to an array, only then is that array copied to that thread’s memory, and only the page of memory (4096 Bytes) that has been changed. This is called a copy-on-write method and is handled by the operating system. Forking is very efficient in this way, only doing the work it needs. For Python, this gets around the Global Interface Lock which is designed to protect python objects. Unfortunately the Windows operating system itself does not have support for the fork function so you cannot run multi-threaded Python codes like pymp on Windows, at least from the Power Shell. However, if you have the Windows Subsystem for Linux (WSL) installed this provides a robust Linux system that bypasses Windows and its limitations so pymp codes can be run in this manner.
The figure below illustrates how multi-threading works on a dot product between two vectors. Since the program uses shared-memory, the initialization is done entirely on the main thread of processor 1. Then each of 4 threads in this example does a partial sum on the vector elements it is responsible for, so all 4 threads are running at the same time but operating on different parts of the data. After they have all completed their parts of the computation, the master thread sums all the partial sums into the dot product and prints it out.
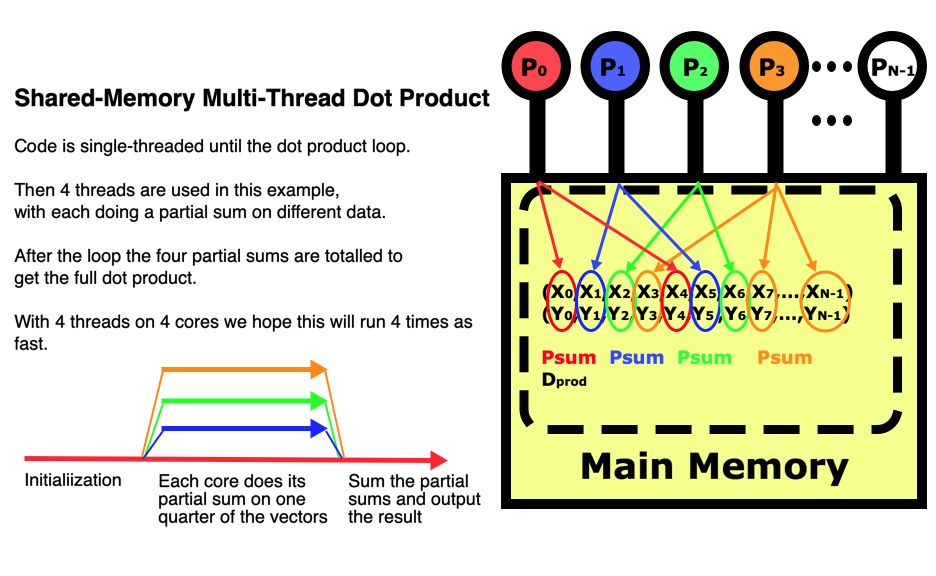
The multi-threaded dot product code
So parallelizing this program really only requires us to change around 11 lines of code, and from that we get the benefit of being able to apply much greater computing power. In Python for example we do have some control over how the parallelization works internally. Using p.range(N) in our for loop will use static scheduling where each thread is responsible for a pre-determined set of indices at regular intervals as in the figure above. If instead we use p.xrange(N) then dynamic scheduling will be used where each index will be assigned to the next available thread. This can be very useful if the amount of work in each pass through the loop varies greatly. Dynamic scheduling can produce much more efficient results in cases where there is a great load imbalance.
Understanding what can cause inefficient scaling
A scaling study is designed to expose inefficiencies in a parallel code and to determine how many cores to use for a given problem size. That last part is important to understand. If there is too little work in each iteration of a loop, then loop overhead can limit scaling. Calculations on larger data sets usually scale better.
A loop may be very scalable in itself, but if there is too much time spent in the scalar part of the code like initialization, doing the reductions, or doing input at the beginning and output at the end, then the entire code may not scale well. Load imbalance can also be a problem. If the time it takes to pass through a loop varies, then using dynamic scheduling is very important.
Shared arrays are an extremely important part of multi-threaded packages. Since they do involve the copy-on-write mechanism, they can lead to inefficiency in the loop. In general this is minimal but something to be aware of.
Multi-threading packages like OpenMP and pymp provide mechanisms that force loops in the algorithm out of multi-threaded operation and back into single-threaded operation. This always leads to terrible scaling and should almost never be used.
Scaling Study of the Multi-Threaded Dot Product Code
- Multi-threaded computing is powerful and fairly easy to use but only works on one compute node.
- Understand key factors that can limit the efficient scaling of multi-threaded programs.
Content from Message-Passing Programs
Last updated on 2025-09-17 | Edit this page
Estimated time: 30 minutes
Overview
Questions
- What is the distributed-memory programming model?
Objectives
- Learn about message-passing in distributed-memory computing.
- Understand the strengths and limitations of this approach.
The Message-Passing Paradigm
While multi-threaded parallelization is very efficient when running on a single compute node, at times we need to apply even more compute power to a single job. Since this will involve more than one compute node, we will need a separate but identical program running on each computer which leads us to a new programming paradigm. This is known by various descriptive names including MPMD or Multiple Program Multiple Data, but is more commonly known as message-passing which is how data is exchanged between multiple identical copies of a program that each operate on different data. There is a common syntax used which is MPI or the Message-Passing Initiative standard. C/C++ and Fortran have several MPI implementations including free OpenMPI and MPICH libraries and commercial Intel MPI, while Python uses mpi4py which implements a stripped down version of the MPI standard. R does not have any message-passing implementation though there was work on Rmpi in the past that was never completed.
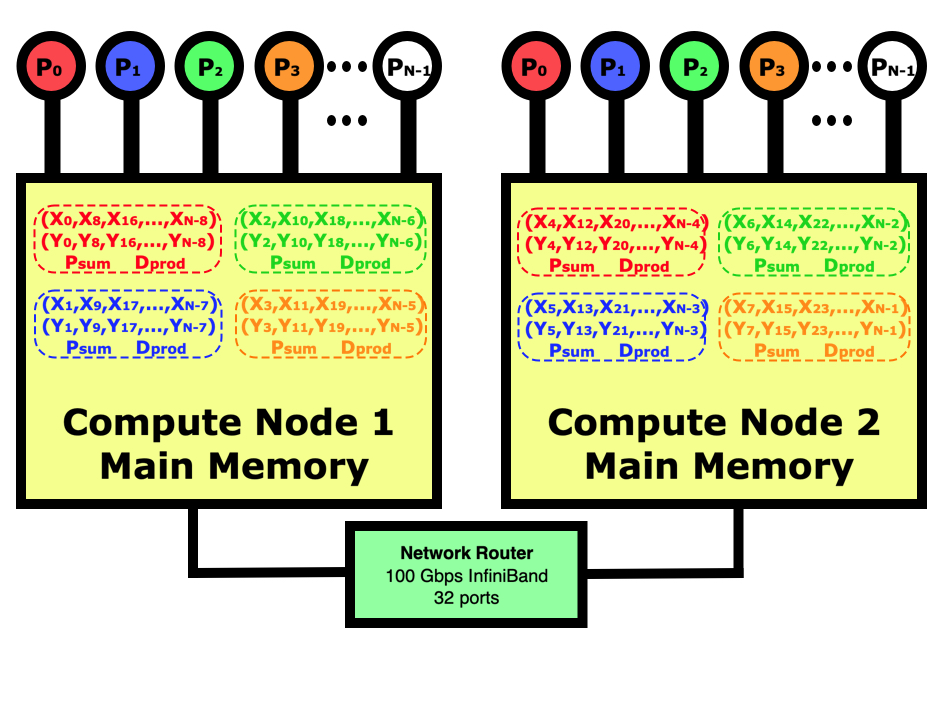
The diagram below shows what a distributed-memory dot product looks like on a multi-node computer in contrast to the shared-memory program in the diagram in the previous chapter. In this case, our job is running on 4 cores on node 1 and 4 cores on node 2. Distributed-memory means that there will be 8 identical programs running, with 4 on each node, but each will have responsibility for doing one eighth of the calculations. We will need to use the mpirun or mpiexec commands to launch eight copies of the program on the two nodes then start them running. The jobs will handshake then decide which part of the data each is responsible for. After all nodes have calculated their partial sums, they will be globally summed across all 8 tasks using the network if needed then the program with lowest rank will print out the results.
In parallel computing, the programmer must decide how to divide the data between the processes. In this case, we could have just as easily decided that rank 0 is responsible for the first 1/8th of the elements, rank 1 for the next 1/8th, etc. If we were reading in the data and distributing it in blocks to each other process then this would be better since we wouldn’t have to move the data around before sending out each block of data. In this case for the dot product, it simply doesn’t matter. Regardless of how the work is divided, it sometimes does not come out evenly so you do have to make sure that all work is accounted for even if some processes have to do slightly more.
The message-passing dot product code
Let’s look at how the message-passing version of the code differs from the original scalar version and contrast it to the multi-threaded version. If you are using Python, you first need to pip install mpi4py into your virtual environment then you can import mpi4py as MPI to bring the package into your code. The compiled languages C/C++/Fortran need an #include<mpi.h> to pull in the headers for MPI, then you compile with mpicc or mpifort.
Since a message-passing job is many identical copies of the same program working on different data, we need to use the mpirun -np 4 command for example to launch 4 copies of the code. If you are running the job using a scheduler like Slur, this will run on the 4 cores that you requested. If you are not running through using a scheduler, you can specify different compute node names such as mpirun –host node1,node1,node2,node2 to run on 2 cores of node1 and 2 cores of node2. You can also specify a hostfile and number of slots using mpirun –hostfile hostfilename where the host file contains lines having the node name and number of slots on that node (node1 slots=2).
All message-passing programs start with an initialization routine which for Python is the comm = MPI.COMM_WORLD statement and C/C++/Fortran is MPI_COMM_WORLD. This says that our communicator includes all ranks available (COMM_WORLD), and it connects with all the other programs. The other two lines that are at the start of every message-passing program are functions to get the number of ranks, the message-passing word for threads, and the rank for each program which ranges from 0 to the number of ranks minus 1. This rank is what the programmer uses to decide which data each copy of the program will work on, and is also used to identify which copy of the program to pass messages to.
Each rank is responsible for doing the dot product on part of the data, and the programmer must decide on how to divide this work up. For this code, we are going to divide the work up in a similar way to how the multi-threaded program worked, where the rank 0 is responsible for indices 0, nranks, 2nranks, 3nranks, etc. The initialization of the X and Y vectors shows how we are now just dealing with N_elements each (N/nranks) but we still want the initialization to be the same so we have to change that a bit.
We do a barrier command before starting the timer so that all the ranks are synchronized. Normally it is good practice to avoid barriers in codes, but in our case we are doing it so we get a more accurate timing.
Each rank calculates a partial sum of the indices that it is responsible for. Then all ranks must participate in a reduction to globally sum the partial sums. Notice at the end that we only have the lowest print its results. If we didn’t protect the print statements like this, we would get nranks copies of each print statement.
If you want to try other examples of MPI code, the scalar algorithm for a matrix multiply is very simple but you can compare that to the MPI versions in the links below. These algorithms are much more complicated since you need to have particular columns and rows in the same rank in order to perform the vector operations. This involves a great deal of communication and the programmer must determine the optimal way to handle the message passing to minimize the communication costs and memory usage.
We see from all this that parallelizing a program using message-passing requires much more work. This is especially true in more complex programs where often you need to read input in on the lowest rank and broadcast the data out to the other ranks. Most of the time there is also a lot of communication needed during a calculation that requires sending data from one rank to the other. In these cases, there must be a pair of send and receive statements to specify the starting data, what node to send it to, and on the receiving rank you must specify the source rank and where to put the data. The nice thing about message-passing is that the libraries do all the work of interacting with the underlying communication system for you.
So while message-passing is more difficult to program, the supporting libraries are very sophisticated in simplifying the process. With multi-threading, your algorithm is limited to the number of cores on a single compute node, but the message-passing model has no limits. People have run message-passing codes on millions of cores on supercomputers worth as much as half a billion dollars.
Understanding what can cause inefficient scaling
In many practical message-passing codes, we would need to read data into one of the ranks, divide the data up and send it out to each other node. This extra communication is necessary but does lead to some inefficiency to be aware of. Real message-passing applications also usually require communication mixed in with the computational part in order to move data to where it needs to be for the calculations. Advanced programmers can try to minimize this by hiding the communications behind the calculations using asynchronous communication calls. All this communication can slow down the job, and this usually gets worse as you spread a job over more cores, and especially over more compute nodes. It is therefore usually best to keep the processes in a run on as few compute nodes as possible so that most of the communication is being done within a node by memory copies that are faster than sending data between nodes across network cards.
Networks in cluster supercomputers like those typical in universities are usually not uniform. Performance within a switch which may connect 32-40 compute nodes may be very fast, but if your job is spread on different network switches it can be much slower since switches may be connected to each other at much lower aggregate bandwidths. In Slurm you can request that your job be run on just one switch by using –switches=1 but this is not always honored.
All of this does sound more complicated as message-passing programming is definitely more complex than the multi-threaded approach. But if you really need to apply more computing power to your job, it is the only way to go.
Scaling Study of the Distributed-Memory Dot Product Code
- Distributed-memory computing is very flexible, extremely scalable, but more difficult to program.
- Understand key factors that can limit the efficient scaling of message-passing programs.
Content from Language Survey
Last updated on 2024-08-13 | Edit this page
Estimated time: 10 minutes
Overview
Questions
- What are the strengths and weaknesses of each computer language?
Objectives
- Understand the criteria we will use to evaluate each language.
People choose a programming language for a given project for many reasons. Sometimes they choose a language because it is the only one they know, or because it is the only one their advisor knows. Many languages are easier to learn and use, and some can be used interactively.
In the next few sections, we want to do a survey of some of the more common languages being used in science so that we can compare and contrast each. When we first choose a language to use for a project, it is common to only consider the capabilities of that language. For example, R has great access to statistical analysis routines so it is a great choice when those capabilities are needed. R and Python both can be used interactively which appeals to many. But when we start talking needing performance as well, then we have to balance the capabilities that each language offers with the need to get great performance. This performance can come in the form of scalar or single-core performance, but also involves the ability to apply multiple cores or multiple compute nodes.
Capability, ease of programming, performance, and parallelizability are all attributes that we will need to consider. Capability refers to the routines each language has access to like all the statistical functions in R, the wide variety of artificial intelligence packages programmed in Python, and the mathematical toolboxes in Matlab. Usability means the ease of programming and the productivity of the programmer. A low level language like C is incredibly flexible and efficient but is more difficult to program and debug so that program development takes longer. Performance is unimportant for simple calculations but everything as we scale up to more complex and computationally costly runs. This is why people may start a project in a less efficient language and end up needing to switch languages when performance begins to limit the science that can be done. Parallelizability refers to how many compute cores we can apply to a given job. This again ultimately limits the size of the science we can achieve. We must understand how each language measures up for each of these merits in order to choose an effective approach for each project we are interested in.
- Capability - Access to the routines and data structures you need
- Usability - Ease of programming and productivity
- Performance - How fast is the final code going to run?
- Parallelizability - How many cores or compute nodes can be used?
Compute cycles on NERSC (National Energy Research Scientific Computing) supercomputers are dominated by the compiled languages C/C++ and Fortran. Python is involved in one quarter of all jobs, but in a job control role rather than a computational one. When you run jobs on large $100 million supercomputers, you have to choose your language for performance reasons even if that means putting extra effort into the programming.
In a university environment it is very common to have less efficient languages supported for computations such as Python, R, and Matlab. Even though these are far from efficient computationally, they are typically easier to program and can provide greater functionality. These factors are often more important in cases where you may have a single programmer writing a custom code for a particular project. So the choice of a language can depend on the circumstances which may include factors like how long an application is expected to be used versus how much effort it will take to be developed.
It is useful to know a little bit about each language so you can decide which is best for a given project or even which languages you want to be proficient at for your career. The next sections will present the strengths and weaknesses of many languages commonly used in scientific computing. Some languages have common practices that are performance bottlenecks that need to be avoided, so these will be discussed and alternative approaches presented.
Overview
C/C++ and Python are row-major languages with arrays starting at 0.
Fortran, R, and Matlab are column-major languages with arrays starting at 1.
C/C++ and Fortran are compiled languages for high-performance.
Python, R, and Matlab have some optimized libraries to help with performance.
- Performance is just one criteria we need to understand when choosing the best language for a given project.
Content from C and C++ Languages
Last updated on 2024-08-13 | Edit this page
Estimated time: 30 minutes
Overview
Questions
- What are the strengths and weaknesses of programming in C and C++?
Objectives
- Analyze the merits of the C/C++ language.
- Learn how to compile and run C/C++ programs.
- Try compiling and running OpenMP and MPI versions.
The C Programming Language
C is a very low level language that is extremely flexible and efficient. It is the language used to program the Linux operating system and the Python, Matlab, and much of the R languages. But all this power comes at a price; it is more difficult to debug.
C files end in .c with header files ending in .h. It is a row-major language meaning that a matrix is stored by rows with elements of each row next to each other. Arrays are numbered starting with zero same as with Python.
In C, variable types are less strict to allow for greater flexibility, but this makes it more difficult for compilers to catch errors before run time. Memory is dynamically allocated in a raw form and assigned with a pointer to the first element, but there is little control after that on how the programmer accesses the memory. If the program tries to write to memory past what is allocated to that array, there is no protection to prevent it from happening. So the programmer is responsible for a lot more since the compiler cannot check much of the code and provide detailed warnings. This is just the cost of the low level access and flexibility of C.
Part of the power of C as well as C++ and Fortran is the access to massive numbers of highly-optimized libraries of routines that have been developed over the past 60 years. These involve scalar, vector, and matrix algorithms in the BLAS (Basic Linear Algebra Subroutines) libraries, sparse matrix libraries, Linear Algebra Package of LaPack and its multi-processor version ScaLapack, FFT (Fast Fourier Analysis) routines, and many others.
OpenMP is the premier multi-threaded package available for scientific computing. There are other methods of doing multi-threaded computing like pThreads that are just as efficient but harder to use. MPI is likewise developed specifically for C/C++ and Fortran. While other languages have stripped down versions implemented, none can rival the rich set of functionality that the OpenMPI and MPICH packages provide with the full MPI standard.
C doesn’t have as much access to statistical package that have been developed for R, nor the mathematical toolboxes of Matlab and the wide variety of artificial intelligence toolkits of Python. But it is unrivaled in power, performance and flexibility.
The C++ Language
C++ is a super set of C, meaning that it starts with C and adds functionality beyond. Since C can be embedded with C++ code you get the best of both worlds with access to the low level capabilities of C along with the high level data structures and programming functionality of C++. C++ files end with .cpp and use the same header files ending in .h.
It is an object-oriented language, with objects having data that can be private (hidden) or public (exposed) along with definitions of how that object is created and interacts with other objects. This is good in a sense since much of the work in creating an object is hidden from the programmer, but hiding this process also means it is more difficult to track memory usage and computations, both of which are very important in understanding performance issues.
In C++ you also have overloaded operators, meaning that a multiplication sign can have different meaning depending on the data types it is applied to. If it is between 2 scalar variables then a scalar multiplication is done, while the same operator between 2 matrices would do a matrix multiplication. This is why C++ is great for programming other languages like R since the programmer can define what each operator does and have that be dependent on the variable types involved.
Much of what makes C++ so powerful is also what makes it more difficult to work with where performance is concerned. The ability to hide object initialization means that memory allocation is also hidden. Operator overloading also can obscure the computations being done, as a multiplication between two variables as in the example code below may represent a single floating-point operation or a triply-nested loop if the operands are both matrices. Memory and computations are simply less explicit in C++ by design, and that can make it more difficult to identify where performance issues may lie.
C/C++ Compilers
Unlike interpreted languages like Python, R, and Matlab, you have to compile C and C++ code into an executable before running. The compiler analyzes the code and optimizes it in ways that cannot be done on the fly with interpreted languages making the resulting executable much more efficient.
The most common compilers are the Gnu compiler gcc with the C++ version g++ and the exceptional commercial Intel compilers icc and icpc. There are many compiler options available but you can’t go wrong using -g to generate a symbol table which will provide a line number where the error occurred in case of a crash and -O3 for high-level optimization. You compile in the OpenMP pragmas using -fopenmp for the Gnu compiles and -openmp for the Intel compilers.
Makefiles for compiling
Compilation is actually done in two stages although for single file applications it is typically done in one step. Source files get compiled into binary object files with a .o suffix then all those are linked together with any optimization libraries to produce the executable.
Large applications may divide the source code into many smaller source files for organizational reasons. A Makefile can be developed that has all the logical directions to compile a single application with a single make command. These dependencies have many advantages, such as speeding up the compilation process by allowing only those source files that have changed to be recompiled into new object files. You will get a chance to examine a small makefile in the exercise at the end of this lesson.
Installing large software packages
To install any large software package you will need to read the documentation and follow the directions. Having said that, many well designed packages follow a similar approach of running a configure script, then compiling the package with make and installing it with make install. The configure script usually requires at least a –prefix= argument to tell it where to install the software. While you will see many variations on this approach it is good to at least understand this as a starting point.
- ./configure –prefix=/path/to/installation/directory
- make
- make install
Practice compiling and running C codes
The most common compiler for C is the Gnu C Compiler gcc. This may be available by default on your system, so try gcc –version to check. If not then you’ll need to figure out how to gain access to it. You can also try the Intel C Compiler icc if you have it available on your system.
Try compiling the dot_product_c.c file using gcc -g -O3 -o dot_product_c dot_product_c.c which tells the compiler to use optimization level 3 and create the executable dot_product_c from the source code dot_product_c.c. Once compiled you can run this using ./dot_product_c or submit a job to the batch queue. Try also to compile with icc if it is available.
Next try to compile the OpenMP multi-threaded version. You will need to tell it to access the OpenMP library using a -fopenmp flag for the gcc compiler or -openmp for icc. Try a few runs with different numbers of threads to get comfortable with running on multiple cores.
If you have an MPI package installed, try compiling the message-passing version using mpicc -g -O3 -o dot_product_c_mpi dot_product_c_mpi.c and running some tests with mpirun -np 4 dot_product_c_mpi for example.
If you want more practice you may try running the matmult_c.c code and the optimized version matmult_cblas.c.
On a modern Intel system the raw scalar code ran in 0.14 seconds as did the single-threaded OpenMP and single-task MPI runs. The test on 4 threads took 0.06 seconds which is quite a bit off the 4x speedup we are looking for. This again is due to how little work is being done during each pass through the loop compared to the loop overhead. The MPI test on 4 tasks is better at 0.047 seconds and is a little faster at 0.034 seconds on 8 tasks since the parallelization is done in a different manner. How do your results compare to these?
- Learn about the characteristics of C/C++
Content from The Fortran Language
Last updated on 2024-08-13 | Edit this page
Estimated time: 20 minutes
Overview
Questions
- What are the strengths and weaknesses of Fortran?
Objectives
- Analyze the merits of Fortran.
- Learn how to compile and run a Fortran program.
- Identify source files for different versions of Fortran.
This Fortran section can certainly be skipped if time is limited. If so, I’d suggest at least saying a few lines from the first paragraph below.
Modern Fortran
Fortran is one of the oldest computer languages used for scientific computing. One reason it is still heavily used is for historic reasons since there are just so many lines of Fortran code out there that are hard to replace. The good news is that this code base is extremely efficient. The Fortran language has also continued to modernize adding much of the same advanced functionality of C++.
Historically people used the f77 or Fortran 77 standard for a long time (defined in 1977). Modern Fortran has made great strides from this old standard in adding object oriented programming capabilities and a less stringent form. Fortran code is identified by the .f suffix or .F if there are pre-processor commands embedded. You may also see the .f90 or .f95 suffix to denote that the code adheres to the fluid formatting of the Fortran 90 and 95 standard defined in 1990 and 1995 respectively. Files with the .mod suffix are modules.
All these newer standards have added capabilities similar to C++ like dynamic memory allocation, object-oriented programming, and operator overloading. More recent work has been geared toward adding more parallel capabilities like the Coarray Fortran parallel execution model and the Do concurrent construct for identifying that a loop has no interdependencies and is therefore capable of being parallelized.
The primary value of Fortran will always be its efficiency and the same access to all the scientific and mathematical packages shared with C/C++. It is a column-major language like R and Matlab, and starts arrays at one instead of zero just like both of those as well. OpenMP and MPI packages likewise have full support for Fortran.
So Fortran is every bit as powerful and efficient as C/C++, but it is slowly being taken over by C/C++ on large supercomputers.
Language characteristics to avoid (gotchas)
While most memory in C/C++ is dynamically allocated, it is very common to have Fortran arrays statically allocated to a given size especially in older codes. This memory comes from what internally is called the stack which is a variable defined on each system. In our cluster at Kansas State University the default stack size as seen by doing ulimit -a is set to only a little over 8 MB while data arrays can easily exceed gigabyte sizes at times. When you exceed the stack size, your job crashes with a segfault that will not give you any useful information on what went wrong. If you think the stack size may be an issue, you can include a command ulimit -s unlimited before running your application to remove the stack size limit entirely.
Compiling Fortran code
Fortran is compiled with many of the same arguments and libraries used for C/C++. The Gnu version is gfortran and the Intel compiler is ifort. When using the OpenMP multi-threading package you will add the -fopenmp or -openmp flag respectively. To compile a Fortran MPI code you will use mpifort.
While there are many compilation options for each of these, you can general get by with -O3 level 3 optimization. I also strongly suggest always compiling with -g. This creates a symbol table so that if your code crashes you at least get the line number where it failed. Without this you get a pretty meaningless onslaught of information that won’t really give you a clue as to the problem. Compiling with -g should not slow down your code as long as you also use -O3, and the extra size of the executable should not matter.
Compiling source code to get an executable is again a two step process where source code is compiled into binary object files which are combined with any libraries needed in the linking stage. For simple applications this may all be done in a single step. For more complex codes involving many individual source files and modules it is common to have a Makefile handle everything. The Makefile provides the logic to compile only the parts of an application code base that have changed, then link everything together to produce the executable.
Practice compiling and running Fortran codes
Try compiling the dot_product_fortran.f90 code with the gfortran compiler, then try with ifort if you have access to it. Do the same with the optimized code dot_product_fortran_opt.f90 to see the difference that the built-in dot_product( x, y ) function can have. You can then compile the OpenMP version dot_product_fortran_openmp.f90 and do a scaling study, and if you are on a system with MPI installed then try compiling and running the MPI version dot_product_fortran_mpi.f90 using mpifort. Once you have compiled these codes manually take a look at the Makefile. This contains all the commands necessary to compile all the codes above with a single command make all_fortran.
Each computer system may be set up differently with regard to what compilers are available and how they need to be accessed. You may need to contact your administrator for help if you are unsure whether you have the Intel compiler suite installed, and how to access an MPI package if available. In my tests on a modern Intel system the raw Fortran code and optimized both took 0.14 seconds as did the OpenMP using 1 thread and the MPI version using 1 task. OpenMP using 4 threads took 0.063 seconds which is a little more than twice as fast and then performance flattened out for more threads. The MPI version using 4 tasks took 0.05 seconds which is slightly better than the OpenMP version, and 0.027 seconds for 8 tasks showing better scaling.
- Learn about the characteristics of modern Fortran
Content from The Python Language
Last updated on 2025-10-07 | Edit this page
Estimated time: 40 minutes
Overview
Questions
- What are the strengths and weaknesses of programming with Python?
Objectives
- Analyze the merits of programming in Python.
- Learn how to work with virtual environments.
- Learn about common performance-oriented libraries.
Python performance
Python is a high-level and extremely flexible language that has broad acceptance within the high-performance computing community and even more broadly with scientific and technical programmers. However, it isn’t a performance language in the same manner as the compiled languages C/C++ and Fortran. It is an interpreted language that executes the code line by line while compiled languages optimize larger blocks of code before execution. At the Department of Energy supercomputing center NERSC Python is involved in 25% of the code bases for the complex applications, but only accounts for 4% of the CPU time on their systems. This means that Python has a high level control role while the compiled languages are used to do the performance computations.
A matrix multiplication algorithm implemented with raw Python code is more than a hundred times slower than raw C code that is compiled. You can achieve decent performance with Python only when your code uses the highly-optimized libraries, and fortunately there is a rich set available such as NumPy and SciPy among many others. Even these can be slower than their compiled language counterparts though, as a NumPy matrix multiplication is still 50%-25% the speed depending on the matrix size compared to a BLAS library DGEMM function available for C/C++/Fortran.
While Python is not intended to be a performance language, there are very good options available for parallelization. The pymp library is a multi-threaded package based on a stripped down version of the much more extensive OpenMP library available for the compiled languages. pymp is actively developed and maintained and provides the basic functionality of OpenMP to the Python environment in an easy to use manner. The mpi4py package likewise provides a stripped down MPI environment for Python users. While neither of these is anywhere near as complex nor complete as their compiled language cousins, both provide the basics that most programmers will need.
Dask is another option that extends Python in two ways. If your data set is too large for the memory on one computer, it provides the framework to use packages like NumPy, Pandas, and Python iterators on data that is distributed across multiple compute nodes in a cluster. It also provides a dynamic task scheduler for parallelizing code.
Numba is designed to speed up raw Python code by compiling blocks of code programmed as functions that are tagged by the programmer with @jit(nopython=True,cache=True). This uses the industry standard LLVM compiler library, but the compilation is not being done once ahead of the runtime as with C/C++ and Fortran, it is being done at runtime in what is referred to as a just-in-time or JIT manner. How effective this approach can be will depend on how much the compilation can be done in advance of when it is needed, and how much optimization it can do on the fly, but there also is the option to save the compiled functions so that they don’t need to be recompiled for subsequent runs. There are also multi-threading options for parallelizing functions.
Parsl is a parallelization library for Python that very easily allows that programmer to define functions as separate apps or applications that can be run on different cores or compute nodes. This approach is especially useful in expressing multi-step workflows. The link below provides more information on this approach.
memory_profiler is a Python package that allows easy profiling of the memory usage of the code. If run externally using the mprof command it reports the full memory usage of the executable which can also be easily plotted. You can also add @profile statements before functions to provide line by line memory profiling to help identify exactly where in your code your large memory allocations are occurring. The link below provides a more detailed explanation with examples.
Python memory-profiler package documentation
The line_profiler package in Python similarly can run externally or internally, but provides timing information in a line-by-line manner. Further information can be found at the link below:
Python capabilities
For what Python lacks in raw performance as an interpreted language it more than makes up for in its flexibility and easy of use. It is common to develop code in an interactive environment like a Jupyter Notebook which provides a very quick cycle from changing code to testing it. It is a high level language that is easy to read, write, and learn, and with advantages like dynamic variable typing. All this makes Python a high productivity language enabling programmers to produce code more quickly than in many lower level, more computationally efficient languages.
Where this language really shines is in the extensive selection of software packages that have been developed for the Python environment. Some important examples are the TensorFlow and Scikit packages for doing Artificial Intelligence and Machine Learning, but Python is commonly used in fields as diverse as video processing, data mining, game and language development, finance, and general programming.
Technical aspects
Python is a row-major language like C/C++ where 2-dimensional arrays or matrices are stored by row with elements in each row being next to each other in memory. Python is also like C/C++ in that arrays are numbered starting with 0. Languages like Fortran, R, and Matlab are the opposite of Python and C in both these respects.
Python is an interpreted language like R and Matlab. It can be compiled but not in the same way as C/C++ and Fortran where the compiler optimizes whole blocks of code. The Python compiler simply packages the code up so that it can run independent of Python or any installed packages. This means the user running the ‘compiled’ python executable does not need access to the same version of Python nor any virtual environment with the python packages installed since everything is packaged up in the executable. There will be an exercise at the end of this section where you will be able to test this for yourself.
Advantages of using a virtual environment
If you are on your own laptop or desktop computer you can install Python packages system wide if you would like. On HPC systems there may be some Python packages installed as loadable modules. You can also install packages locally on your home directory using the –user flag to pip install.
It is strongly advised however that you use a different virtual environment for each new project. While you may need to re-install some common packages like NumPy, having a clean virtual environment for each project helps avoid problems with conflicting versions of dependency packages. Quite often if users are having trouble installing Python software, I’ll advise them to start with a clean virtual environment and that fixes everything.
Creating a virtual environment is not very difficult. Below is an example of creating one called env-py-3.7.4 where I labeled it after the version of Python being used. Once activated the prompt will change to show that you are working in the env-py-3.7.4 environment. You can install any packages you need, run your python code, then deactivate the environment when you are done. The dollar sign is the command prompt to illustrate how the prompt changes while the virtual environment is active.
Compiling Python
Compiling Python code does not do the same type of code loop analysis that compiling C/C++ or Fortran does, so there really isn’t any speedup involved. It does create a self-contained executable that no longer needs to have a matching Python version or installed package virtual environment available. It is therefore most useful when you are done developing the code and want to distribute it as a binary.
Compiling Python code does vary depending on the operating system you are on. In Linux you can compile code by adding a flag -m py_compile which will create an executable in a pycache directory.
BASH
python -m py_compile python_code.py
mv __pycache__/python_code.cpython-37.pyc compiled_python_code.pycThe executable compiled_python_code.pyc will then run on systems where Python is not installed and you have no virtual environment activated.
Language characteristics to be aware of (gotchas)
One of Python’s greatest strengths is that it is a simple high-level language that is interpreted and easy to use. This is also its biggest disadvantage when it comes to performance. Simply put, when you need performance you need to use the highly-optimized numeric libraries. Unfortunately compiling Python code really doesn’t help when there are no optimized library codes available.
Virtual environments provide an excellent way to manage the installation of software packages, and it is easy to use the pip install command which installs the desired package and any dependencies. You do always need to have the active virtual environment match the Python version number used to install all those packages, so if you want to use a newer version of Python you will have to reinstall all the packages too.
Python2 was deprecated on January 1 of 2020. Everyone should be programming in Python3 now, and these codes are not backwards compatible with Python2. This means that if you encounter any Python2 code you would either need to convert the code to Python3 or find an old and deprecated version of Python2 and hope that still works. Basically if you run into old python code at this point, expect to have some errors to deal with.
Python uses a Global Interpreter Lock (GIL) to ensure that only one thread can control the interpreter at any given time. This basically defines Python as being single-threaded which simplifies everything internally. The problem comes when a programmer wants to do multi-threading to apply more power to solve a problem faster. The pymp package gets around the GIL by forking off separate processes and therefore each essentially has its own GIL. The mpi4py package for message-passing is not affected since with MPI each task is a separate copy of the same program. In general, the GIL is something to be aware of but packages like pymp have already done the work of getting around the issue.
Play with the Python dot product examples
Take a look at the Python versions of the dot product code to see how they differ from the C and Fortran versions. Also compare the pymp multi-threaded and mpi4py message-passing versions to the scalar version to see what changes were made. If you haven’t already, run the scalar code and do scaling studies with the multi-threaded and message-passing versions to see how they compare with versions written in other languages.
While the syntax is different, the code is basically the same in each language. Performance is much slower for raw Python code than the compiled language. From looking at the same code in various languages, which do you think would be easiest to write from scratch?
Compare the raw matrix multiply code to a NumPy version
Do a scaling study of the matmult.py code for various matrix sizes of 10, 100, and 1000. Compare this to the matmult_numpy.py version to see how much of a difference the highly-tuned library function makes.
If you have already run the optimized C version, you may notice that the compiled code still beats the optimized NumPy routine by 2-4 times in my tests. Using the NumPy routine closes the performance gap enormously but the C version is still better.
Optional Exercise - Try compiling a Python code
If you have time, try compiling one of the Python test codes supplied like matmult_numpy.py. Then try running it without an active virtual environment meaning that there is no NumPy library installed.
Once compiled you should be able to run this with a command like: ./matmult.pyc 100.
Optional Exercise - Try implementing memory-profiler or line_profiler
If you have time, try implementing memory-profiler or line_profiler in one of the Python test codes supplied like matmult_numpy.py. Then try running the code to see the memory or timing output.
Optional Homework - Test the Numba version of the dot product code
Do a pip install numba then time the dot_product_numba.py performance and compare to the raw code. Does the performance change if you cache the compilation by adding cache=True? You will need to run twice so that the second time takes advantage of the cached compilation code.
Notice that in this code we needed to rewrite the computationally intensive loops into functions. This does not take much effort but does disrupt the program flow somewhat, but if it speeds up the runtime it is usually worth it. Unfortunately in the case of the dot product the Numba version takes 13 seconds compared to only 90 milliseconds for the raw code. Adding cache=True to cache the compiled code does reduce the runtime down to 3.7 seconds but this is still substantially worse than the original code. There are also warnings about lists being deprecated as input arguments in the near future. Switching our algorithm to use NumPy arrays may be necessary but this also defeats the purpose of using Numba since NumPy already has optimized routines for the algorithms we are testing. So in short this isn’t a really good test of the capabilities of Numba, but is a good example of how these things don’t always work as hoped, and it illustrates that Numba does not support all aspects of Python. Forcing the computational parts of an algorithm into functions is also detracts from the flow of any program, and is definitely not what is considered as the Python way.
Optional Homework - Write a Pi calculation program using Numba
Write a Python version of the Pi calculation program and time raw Python code versus Numba optimized. If you want to have more fun try out Numba multi-threading compared to pymp for this same algorithm. This is a much fairer test of the capabilities of Numba since we are not comparing it to optimized functions in NumPy.
If you do this please contribute your code and timings.
- Learn about the characteristics of the Python language.
- When performance is important always use optimized libraries!!!
Content from The R Language
Last updated on 2025-09-12 | Edit this page
Estimated time: 50 minutes
Overview
Questions
- What are the strengths and weaknesses of the R programming language?
Objectives
- Analyze the merits of the R Language.
- Learn what to avoid in R when performance is important.
- Learn a little about how to parallelize R code.
Programming in the R Language
R is a high level programming language with an extremely rich set of internal functions and add-on packages for statistical analysis and other scientific programming. It is an interpretive language and therefore the raw performance is not great. While there are ways to write R code that performs better, it does take more work than in other languages due to the many performance pitfalls inherent in the language, and often you have to sift through the the many external packages to find ones that work well for your needs. This section will try to identify those pitfalls and present higher-performing alternatives.
R code is identified with the .R ending. External add-on packages are installed into the user’s base directory under a sub-directory also named R. These packages can be easily installed from the CRAN mirrors using commands like install.packages(“data.table”) then referenced in the code with a similar statement like library(data.table). The wide range of scientific and code packages available in R and their ease of installation and use are the language’s real strengths.
R programs can be run through the Linux command line interface (CLI), submitted to batch queues, or run interactively from the CLI or the popular graphical user interface Rstudio. The package Rshiny also makes it easy to build interactive web applications from R.
R is like Python in that both are interpretive languages and both can easily be used interactively which is helpful for minimizing the code development cycle. The programmer is constantly changing the code and immediately seeing the results. This strength in easing the development cycle is also one source of its weaknesses in performance as interpretive languages only execute a single line of code at a time while compiled languages take whole blocks of code like the bodies of loops and highly optimize them before execution time, but this is the same trade-off that many languages face.
You can run R codes on multiple cores, but the parallel capabilities of R are much more limited than the compiled languages C/C++ and Fortran and even compared to Python. The basic parallel model for R is that loops whether from lapply() or foreach return an object that is either a list or a data frame with one entry for the results from each pass through the loop. If your code uses the loops in this manner then parallelizing them is relatively easy as you just need to choose a back-end package to use, tell it how many cores you want to use, then change the lapply() to an mclapply() or the %do% in the foreach to a %%dopar%.
There are many multi-threaded back end options to choose from. The parallel library is now built into R and is a merger of the multicore and snow libraries. The dot_product_threaded.R code shows that the programming approach is a little clunky in that the body of the loop needs to be put in a function, but the efficiency is comparable to other back ends. The doParallel library is a merger of the doSNOW and doMC libraries. The foreach command with dopar make for cleaner programming that looks more like traditional loops. The dot_product_threaded_dopar.R code shows that a little extra programming can be used to greatly reduced the overhead even for loops with little computatotions.
However, it is very common in scientific codes to want to instead have all parallel tasks operate on a shared-memory object like the resulting matrix in our matrix multiplication example. People in the R community often refer to back-end methods that use a fork as shared-memory since the data structures can be referenced in place in a shared manner as long as they are not written to. This is quite different than what the HPC community refers to as shared-memory, where all threads can write to a common data structure and it is up to the programmer to ensure that threads don’t obstruct each other.
Our matrix multiplication example illustrates this difference as the source matrices A and B can be shared in R since they are read-only, saving great time when a back-end that uses a virtual-memory fork is used, but the elements of the result matrix C needs to be filled in by each thread. In R this is very difficult while it is common place in the compiled languages C/C++/Fortran as well as in Python. I have not found any example code yet to do this in R though I have seen references that it can be done with a bigmemory package and a lot of jumping through hoops that goes well beyond the scope of this course. There are also some methods of altering the default matrix multiplication package used in the **%*%** operation but these are very operating-system dependent.
There is extensive support for running C/C++/Fortran code across multiple nodes using the message-passing interface MPI, and Python has a stripped down version of this with the mpi4py package. There is a package for R called Rmpi that provides wrappers around some of the common MPI functions, but it old code that is being updated every few years and it can be difficult to get working as you need certain versions of R and OpenMPI for everything to work. Under Windows you must compile with Microsoft MPI, and there are some limitations in functionality.
The doMPI back-end to foreach runs on this Rmpi package. This perfectly fits into the R parallelization model where the programmer can just slip in a different back end to fit their needs, but this often results in poor parallel performance which is the case with doMPI. The dot_product_doMPI.R code can be used to test the performance for a simple application compared multi-core back ends. There’s also a newer package called Rhpc which supposedly provides better performance but it was removed from the CRAN repository some years ago.
There is a newer package called pdbMPI where the pbd stands for Programming with Big Data. This is an interface to the Message-Passing Initiative that is the foundation for distributed parallel computing in the HPC community, and also is a dependency for the pdbDMAT and kazaam packages for working with matrices across multiple compute nodes. These packages are much more recently developed and while still having 0.x version numbers they are actively managed and being used on large supercomputer systems. These provide true interfaces to MPI functions, not a back-end to mclapply() or dopar, so they are more difficult to use but also much more powerful.
Performance Pitfalls in R
While all languages take effort to optimize and parallelize when performance becomes important, with R it is often more about what aspects of the language need to be avoided that are inhibiting performance.
Profiling function
As with most languages there are many methods that can be used to time sections of code in order to understand where time is being spent. For R the best options are to bracket the code of interest with calls like t_start <- proc.time()[[3]] and t_end <- proc.time()[[3]] then take the difference. The proc.time() function will provide the best clock available and the [[3]] part takes the elapsed or real time that we want. The system.time() function can also be used which returns the time taken to execute the function put in the parentheses. This uses the same proc.time() clock but may provide a more convenient method in some cases.
Another common approach that should be avoided is to use the Sys.time() function. This similarly reports the time between the bracketed code, but it by default auto-adjusts the units to the length of the interval. So if your code takes 59 seconds it will report 59, but if the same code takes 60 seconds it will auto-adjust to minutes and report 1 instead. You can and always should manually specify the units if you choose to use this function.
Dataframes and the rbind() function
Dataframes are a very valuable and integral part of the R language. The results from loops or functions are often returned in the form of dataframes, and the input and output of data is built more on dumping out whole dataframes to files than the line-by-line approaches that other languages use. Dataframes in R are designed internally to be very flexible to enable all of this, but this same design choice makes them extremely inefficient from a computational view when working with larger data sets.
The best example of this is the rbind() function which is used to build a dataframe. It is very common to build a row of data using cbind()then use rbind() to add the row to the dataframe table, but internally R must allocate an entirely new area of memory and copy all the existing data over as well as the new data. This is because R is a column-major language so elements in a column are stored next to each other. If R was row-major then other alternatives would be present like having an array pointing to each row in memory.
Having to recopy the entire dataframe each time a row is added is an enormous performance penalty. I was approached with an R code and asked to optimize and parallelize it since it was going to take a month of runtime to complete. We generated a test case that took one hour, and after commenting out only the rbind() function the calculations took only 5 seconds. All the rest of the time was spent copying the dataframe data to newly allocated memory each time a row was added to the bottom.
If the code is building the dataframe just to dump it out to a file, then one option is to simply print each row to file. R isn’t really designed as well for this so it isn’t always optimal, but often is a huge improvement over the rbind() inefficiency.
A better option is to use the data.table package which is a drop in alternative to a dataframe. It is not as easy to use, but is immensely more efficient than a dataframe since you can pre-allocate the structure and insert values rather than having to constantly rebuild the dataframe structure. There will be an exercise at the end of this section that will allow you to see the difference in the code and measure the performance of each approach.
Parallelizing R code
As mentioned above, when a loop is parallelizable it is conceptually fairly easy to accomplish this. The lapply() function has a multi-core version mclapply() that spawns multiple threads on a single compute node. The foreach command can be parallelized by changing the %do% to %dopar%. Both of these commands are part of the core parallel library in R, but to make them work you need to decide which of the many back-end library packages to use.
Considerations include whether you may need to run on Windows OS which does not support the fork() function that is the more efficient way to implement the needed functionality, whether you may need to run a job across multiple compute nodes, and whether you need to use shared-memory in your threads to enable working on a common data set. Packages based on the fork() mechanism use virtual memory rather than redundantly copying all data structures needed. This can be enormously more efficient when dealing with large data structures as each thread only gets a copy of the pages in memory that it needs to alter. In our matrix multiplication example, that means that the two matrices that only need to be read never get copied to each thread, and the for matrix that is being calculated only the parts that each thread is modifying get copied. For this reason it is always recommended to use a back-end library based on fork(), but Windows does not support this functionality so you may need to consider other options that fully copy all data structures at the start or even a socket-based cluster if you think your code might need to run on Windows. Another option would be to install the Windows Subsystem for Linux (WSL) which supports the fork) function.
The basic parallelization model for both approaches is the same, to have each pass through the loop executed on different cores with one line of data being returned for each iteration in the form of a list or data frame. When the goal is to instead operate on a common data set such as in our matrix multiplication example, then shared-memory is needed. There are not very many back-end packages that support this approach even though it is a very common need. In our matrix multiplication example code we use the mcparallel back-end and the bigmemory package. These are designed to work on matrices, but would not work if you for example wanted all threads to work on a different data structure like a shared-memory data.table.
mclapply() pitfalls
The mclapply() function is fairly straight forward to use since you mostly need to supply the number of cores through the mc.cores= argument. There are options to tune the way the parallelization is done. The mc.preschedule=mRUE argument is the default, and this means that the number of iterations is divided among the available cores at the start. This is highly recommended since if this is turned off the system will fork a new process for each iteration, do the work, then collapse that fork. This can be incredibly inefficient since it means copying data structures many times over so it should in general be avoided. If you do try this, make sure to check the performance for both options as this choice can drastically effect the efficiency of the resulting parallel implementation.
Compare raw and optimized performance of matmult.R
Run matmult_loops.R, matmult_foreach.R, and matmult_builtin.R for matrix sizes 100 and 1000 to compare the performance of a raw loop to the built-in matrix multiplication function. Also compare these numbers to other languages.
I measured 500 seconds for the raw loop in R and 0.10 seconds for the optimized built-in matrix multiplication function. Be aware that the built-in function may use all the cores it has access to, so this may not be a fair comparison unless you submit a batch scheduler job with only 1 core allocated. Python for comparison took 300 seconds for raw loops and 0.13 seconds for the numpy optimized routine, but a larger run for size 10,000 had numpy at 46 seconds compared to R at 72 seconds. So in general, R and Python are similar in speed for both raw and optimized code, but Python is a little faster.
Test the scaling of the rbind() function
Profile the run time for using rbind() as the number of rows in the data frame increases. Time runs of rbind.R for 10, 100, 1000, and 10000 rows.
While rbind() is convenient and works well for small data frames, the time to add rows begins to increase exponentially for data frames around 10,000 rows. I measured 1 second for 1000 rows, 48 seconds for 10,000 rows, and 5100 seconds for 100,000 rows. rbind() works well for small data frames, but it is very inefficient when you scale up to larger data sets of over 10,000 rows. This is because R copies the entire data frame over each time it adds a new row.
Investigate the data table performance.
Test the datatable.R code for 100, 1000, and 10000 rows and compare to the rbind() results.
For 1,000 rows I measured 0.12 seconds for a data table set() compared to 5.4 seconds for a data table assignment and 1.1 second for a data frame rbind(). For 10,000 rows the performance really starts to differ with 0.53 seconds for a data table set() compared to 50 seconds for a data table assignment and 48 seconds for a data frame rbind(). For a large test of 100,000 rows the data table set() still only took 5 seconds while the data table assignment took 485 seconds and the data frame rbind() took 5100 seconds, or 1000x longer. This again shows that while data frames can be convenient, when you scale up to larger sizes you have to use data tables and the set() function.
Optional Homework - Test the IO performance in R.
Test the fread.R code to see the speedup of the fread() function from the optimized data.table package compared to the standard read.csv(). Time runs of fread.R for 10,000 rows, 100,000 rows, and 1,000,000 rows.
For 10,000 rows I saw similar results for each function, but for 100,000 rows fread() was 10 times faster than read.csv() and for 1,000,000 rows it was 100 times faster. This is another example illustrating when to avoid the core R functionality and use the external add-on packages to achieve performance in your code.
Advanced Homework - Use the pdbMPI package to code and run a parallel Hello World program
For those who want a challenge, follow the pdbMPI link at the end of this lesson an write, run, and test the Hello World program.
I would love to have a pdbMPI-based matrix multiply code available for people to look at and test if anyone finds one.
Summary
R is a very powerful language because of the enormous set of statistical and external scientific libraries that have been developed for it. It can however be difficult to program in since much effort involves programming in supplemental packages rather than the core language. Users need to therefore know not only the core R language, but be familiar with which external programming packages to use when more performance or flexibility is needed, and this can be an ever changing target.
It is often very challenging to get good performance out of R code. Elements of the core language like dataframes have inherent performance and scaling problems. Parallelization seems as easy as registering the desired number of cores and changing the %do% in a foreach loop to %dopar%, but setting up writable shared-memory is difficult to impossible. Each back-end package has different capabilities and efficiencies so it can be difficult to decide which approach is best. While it is possible to achieve good performance with R code, much of the work involves programming around the built-in capabilities using optimized add-on libraries, and you have to understand which of the many packages to utilize. It is hoped that this section can at least steer people in the correct direction with some of these performance oriented issues.
- Learn about the characteristics of the R language.
Content from The Matlab Language
Last updated on 2024-08-13 | Edit this page
Estimated time: 30 minutes
Overview
Questions
- What are the strengths and weaknesses of the Matlab language?
Objectives
- Understand the performance and parallelization characteristics of Matlab.
- Learn the practicality of using the Matlab compiler on HPC systems.
Programming in the Matlab Language
Matlab is a commercial programming language designed for broad use in science and mathematics. The obvious drawback of it being commercial software is that it costs money and has licensing restrictions when you use it. The more positive aspect is that it is professionally developed and maintained with lots of advanced science modules.
The core Matlab code is interpreted which limits its speed but there are lots of highly optimized and parallelized low level operations that often lead to very good performance. Users can also parallelize code manually using parfor loops which are easy to implement but difficult to make efficient. Many modules are also programmed to run on NVIDIA GPUs for acceleration.
There is a free language GNU Octave that runs much of raw Matlab code. Some of the more advanced features are not supported, but some like parallelization simply have a different format. Octave also has lots of user-contributed modules covering a broad range of science and mathematics.
Matlab Toolboxes
The Matlab core can be enhanced with a very wide variety of add-on toolboxes. Some are more generic while others are very specific to certain areas of science. Some of the more common ones include:
- Parallel Computing Toolbox
- Simulink - Simulation and Model-Based Design
- Statistics and Machine Learning
- Curve Fitting
- Control Systems
- Signal Processing
- Mapping
- System Identification
- Deep Learning
- DSP System
- Datafeed
- Financial
- Image Processing
- Text Analytics
- Predictive Maintenance
There are many domain-specific toolboxes that include Bioinformatics, Aerospace, Wavelets, and Econometrics to name a few.
Commercial licensing restrictions and costs
Matlab has a base cost with toolboxes being extra. There are annual and perpetual licenses with all pricing being in U.S. dollars.
There are huge educational discounts with students paying under a hundred U.S. dollars for Matlab, Simulink, and 10 add-on toolboxes. Teachers and researchers also get large discounts, but add-on toolboxes can increase the cost quickly as the more common ones can run a few hundred dollars and more specialized toolboxes even more.
Licenses for HPC clusters can be floating meaning that each license can be checked out by an authorized user and used on a given number of processors or nodes. This does require a license server to be set up which can be done on the HPC cluster or handled remotely. Just be aware that with commercial software there is additional setup required as well as costs.
Using the Matlab compiler
When we talk about compiling for C/C++ and Fortran, we are talking about analyzing a block of code like the body of a loop to optimize the code so that it runs faster. Matlab and Python both have compilers but neither does this. In both cases the compiler packages up the code, interpreter, and libraries into an executable that can be run independently. The compiled code therefore does not run any faster.
A programmer will always start by developing and running their code in raw form which requires checking out a Matlab license. If you need to share your Matlab code with others who do not have access to a Matlab license, then you would want to compile the code to package it into an executable that doesn’t need Matlab to run. In an HPC cluster context, you still want to develop your code in raw Matlab but when it is time to run you want to again compile it into an executable so that many copies can be run without needing a separate Matlab license for each. In this way, an HPC center only needs as many Matlab licenses as they want to have simultaneous users developing code rather than needing to support the number of jobs run.
Performance
Matlab is not a compiled language so raw code is much slower than C/C++/Fortran. It is however faster than Python and R as shown in the serial matrix multiply of 1000x1000 matrices taking 5 seconds compared to Python at 306 seconds. The built-in routines that are optimized and parallelized are going to be similar in the different languages, and in this case Matlab takes 0.7 seconds while the Numpy version in Python takes 0.13 seconds in the same test.
Matlab does have very strong integration with optimized and parallelized library routines throughout its modules which brings automatic efficiency and parallelization when available.
Parallelization methods
The Parallel Computing Toolbox is the source of most of the parallel computing capabilities in Matlab. It provides the ability to program using multiple cores, multiple nodes, and GPUs without explicitly using CUDA or MPI. Matlab also includes a wide variety of parallelized numerical libraries at its core to automatically take advantage of the hardware you allocate to your job. The Simulink toolkit allows the user to set up and run multiple simulations of a model in parallel. The Matlab Parallel Server can also be used to run matrix calculations that are too large to fit in the memory of a single computer.
Parallelizing loops with parfor
When iterations of a for loop do not depend on each other, the iterations can be spread across multiple processes within the same compute node in a distributed-memory manner, multiple threads in a shared-memory approach, or they can be spread across multiple nodes. All 3 approaches are accomplished by changing the for loop to a parfor loop. Computations will be split across multiple cores whether they are on the same compute node or multiple nodes. This provides a very easy means of parallelizing code and the flexibility of running the same code in a variety of parallel environments.
This flexibility comes with a prices though. Distributed-memory approaches are often only efficient when the flow of data between processes is careful controlled which is not possible here. It often requires much more work to get the needed efficiency out of complex algorithms. This type of automatic distributed-memory approach also results in large data sets being redundantly copied to all processes which can lead to extra execution time for the communications and much extra memory usage. So if you are working with a 1 GB size matrix and want to run on a 128 core AMD system you would have to copy and redundantly store 128 GB of data at the start.
The multi-threaded approach is designed to avoid this redundant memory use by leaving read-only data sets in place rather than copying them to each thread. This holds much greater promise conceptually, but tests with a simple parallel matrix multiplication are showing much slower times than expected. Even a parallel dot product which is trivially parallel takes longer than the serial version so it is unclear how useful even the multi-threaded parfor is in general.
So while Matlab provides an easy-to-use and flexible parallel programming environment with parfor, it can suffer greatly when dealing with large data sets and complex algorithms, and doesn’t even do all that well on simple algorithms. From these tests so far I would recommend using this approach mostly for trivially parallel algorithms and smaller data sets and testing very carefully.
NOTE: Loop iterations are non-deterministic and indices must be consecutive increasing integers, and there is no nesting of loops allowed.
Octave
GNU Octave is a language that is largely compatible with Matlab, but it is free and unlicensed. It can be used to run many Matlab programs using octave < matlab_code.m though many advanced features of Matlab may not be supported. There are many add-on packages available for Octave but these are different from those available for Matlab.
There is a parallel toolbox that is well developed. This provides a local parallel execution environment similar to the single-node multi-process capability of parpool. There are also tools to work with clusters of computers, but these are more similar to message-passing commands where you manually send and receive data to and from remote processes and manually initiate function evaluation.
Test the performance of the Matlab matrix multiplication code.
If you have access to a Matlab license or Octave, test the performance of the matmult.m code for a matrix size of 1000 and compare to other languages.
For the 1000x1000 matrices, I measure 5 seconds for the serial code, 0.7 seconds for the built-in matrix multiply that uses low level optimized BLAS routines. The parpool multi-process test takes 530 seconds which is understandably slow since it is doing the matrix multiplication in a distributed memory manner without explicitly programming it to do this efficiently. The multi-threaded parpool test measured in at > 510 seconds which is very disappointing since there should be no copying of the matrices at the beginning. It isn’t clear what is happening behind the scenes for this to be so slow.
Test the performance of the Matlab dot product code.
Test the performance of the dot_product.m code for an array size of 100,000,000 and compare to other languages.
I measure serial performance at 0.5 seconds with the built in optimized routine at 0.2 seconds on 8 cores for a modest speedup. The multi-core parfor loop on the same 8 cores takes 2.1 seconds while the multi-threaded parfor loop takes a disappointing 0.9 seconds which is still greater than the serial code. The overhead for using these methods is still much larger than the performance gain which indicates the parfor method should only really be used for very coarse grained algorithms.
Optional Homework: Alter the matmult.m code to run on multiple nodes and multiple cores.
Test on multiple compute nodes and compare performance to the serial and multi-threaded versions. If you want a real challenge try setting the code up to run on a cloud server. And an even bigger challenge would be to convert matmult.m to run on Octave’s parallel computing environments.
If you do develop code for this, let us know so we can consider including your work for others to see.
Summary
The greatest value of Matlab is the very wide range of professionally developed and maintained packages that are available in many areas of science. This comes at a financial cost that often limits how codes can be used, but some of this can be alleviated in an HPC environment by using the Matlab mcc compiler to create an executable that does not need a license to run on many compute nodes at once.
Matlab code itself is not that fast, but it uses highly-optimized library routines seamlessly whenever possible. Adding parallelism into a code manually is often as easy as changing the for loop to a parfor loop, but flexibility and ease of use often do not produce efficient code. These methods are probably only useful for trivially parallel algorithms.
Octave is a viable option to avoid the cost of Matlab, and has many add-on packages of its own as well as parallel computing capabilities. You should however only expect the more basic Matlab codes to run with Octave, then you would need to choose to split off into the Octave world itself.
- Learn about the characteristics of the Matlab language.
Content from Array Jobs
Last updated on 2024-08-13 | Edit this page
Estimated time: 15 minutes
Overview
Questions
- What do array jobs have to do with high-performance computing?
Objectives
- Learn what an array job is in a batch scheduler.
- Understand what types of science can make use of array jobs.
Array jobs using Slurm
An array job is mostly just like a normal job script in that it has arguments at the top prefaced with #SBATCH that tell the scheduler what resources are being requested followed by a list of commands to be executed at run time. The difference is that array jobs have an extra allocation request line like #SBATCH –array=1-5 that tells the Slurm scheduler to launch in this case 5 individual jobs identical in every way except that each will have a different value for the environmental variable $SLURM_ARRAY_TASK_ID. This variable can be used to make each run unique as part of a series of related runs. It might be used as an input argument to the code being run in order to let the application determine what is different with each run. It may also be used to choose a different input file from a list to use when running the application. This is very flexible and is entirely up to the programmer to decide how to use it.
For the job below the $SLURM_ARRAY_TASK_ID variable will be set to 1, 3, or 5. since the script specifies that the task ID starts at 1, ends at 5, and steps by 2.
BASH
#!/bin/bash -l
#SBATCH --job-name=array_test
#SBATCH --time=0-0:1:00
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --mem=1G
#SBATCH --array=1-5:2
hostname
echo "Hello from array task ID $SLURM_ARRAY_TASK_ID"Submit an array job
If you have access to an HPC system with Slurm installed, submit the sb.array_test script from the scripts sub-directory using sbatch sb.array_test then look at the output. If your HPC system has a different scheduler you may need to figure out how to submit an array job yourself and will need to write the job script from scratch. If you want more of a challenge you can try writing a script to choose an input filename from a list using $SLURM_ARRAY_TASK_ID.
This will run 3 individual jobs that will show the $SLURM_ARRAY_TASK_ID to be 1, 3, or 5.
Array jobs as parallel computing
Most of the time when we thing of parallel computing we think of taking a single program and making it run faster by applying more compute power to it in terms of more compute cores. Array jobs allow us to do the same thing, but in this case we are running many individual programs instead of just one.
One common area of science where we can make use of this is to do what is called a parameter sweep. You may have a set of parameters such as system temperature, pressure, atom type, and lattice type (atomic arrangement) where you need to run the same code on all these different input parameters. Array jobs allow you to do parameter sweeps like this in a very convenient manner with a single job script. This makes it easy to submit and manage.
Another common use is in doing statistical science. For applications that use a random number sequence, you may want to run the same simulation many times using a different seed to determine how the results vary statistically as the random number sequence changes.
Programming habits to avoid
Many programmers write scripts to submit lots of individual jobs instead of making use of the array jobs functionality. While the result is basically the same, this method should be avoided in general. Lots of individual jobs can clog up the queue making it difficult for users to see where other jobs are, and can also affect scheduling since batch systems have limits on how deep they can look. Array jobs avoid both of these issues and make it much easier to manage the resulting jobs since canceling your array job is for instance just the same as canceling an individual job.
The testing cycle is always more important when you are dealing with large numbers of jobs. One user submitted an array job for tens of thousands of tasks that had a typo in the email address so when it ran it spammed the ticket system with tens of thousands of bounced emails. Always start by running a few typical jobs to nail down your resource requests. Running a test job with just a few array IDs will allow you to ensure that each job is using the $SLURM_ARRAY_TASK_ID in the desired manner. Then when you are confident that your script is working as intended you are ready to submit the full array job.
- Array jobs provide an easy way submit and manage large numbers of similar jobs.
- Array jobs are another way to do parallel computing, but by running lots of small jobs individually.
- Test your script carefully on a few array IDs before submitting the full job.
Content from Accelerating Scientific Computing with GPUs
Last updated on 2024-08-13 | Edit this page
Estimated time: 15 minutes
Overview
Questions
- What differences are there in handling applications that can use a GPU?
Objectives
- Learn a little about how GPUs can accelerate some scientific codes.
- Understand the basics of compiling and running GPU codes.
How to use a GPU to Accelerate Scientific Calculations?
We normally think of a Graphics Processing Unit or GPU in terms of displaying graphics to a computer monitor. GPU cards are designed with many streaming processors that are great at performing lots of similar computations. While this makes them ideal for doing graphics work, it also makes them good for running some scientific codes. GPU cards can accelerate some scientific codes by an order of magnitude while costing much less than what the equivalent CPUs would.
NVIDIA dominates the GPU accelerating market with both 32-bit or consumer grade GPUs like the NVIDIA RTX 4090 that can cost around $1500, but they also produce 64-bit Tesla cards that have no graphics ports at all and are only designed for scientific computing. These include the NVIDIA A100 which can cost as much as $11,000 each or their newest H100 that can cost twice that. Compute nodes can host 1-8 GPU cards each and in some systems the high-end A100s and H100s may be linked together through a fast NVLink connection. Otherwise a multi-GPU run would need to communication across the PCI bus.
AMD GPUs can also be used to accelerate scientific jobs and are increasingly being found in the fastest supercomputers in the world. The code development is still behind that of the NVIDIA CUDA community due to its much later entrance into the scientific computing market. AMD has a Hip compiler that is used to generate code for the AMD GPUs, and it has the benefit of being able to compile the same code for NVIDIA GPUs as well. The AMD GPU line includes 32-bit GPUs such as the Radeon RX 6000 series and the 64-bit Radeon Instinct MI100 cards with costs comparable to the NVIDIA line.
Using GPUs for Machine Learning
Artificial Intelligence applications like Machine Learning can be done on any CPU or GPU. However, GPUs with tensor units can do them faster and less costly since tensor units can do more operations per clock cycle, and the lower precision of the results do not matter for Machine Learning applications. If your system is going to be primarily used for Machine Learning, you’ll want to look at the tensor units in the GPUs while the memory will limit the size of the system you can work with.
AMD and Intel GPUs do not have tensor units. NVIDIA offers tensor units in their 32-bit RTX GPUs providing an inexpensive means of accelerating machine learning algorithms. The 32-bit NVIDIA RTX 3090 for example offers 328 Tensor cores along with 10496 CUDA cores with a maximum of 24 GB of memory. The 64-bit NVIDIA A100 offers 432 Tensor cores along with 6912 CUDA cores for high-end computing and 40 GB or 80 GB of memory. These A100s can also be combined with the NVLink connection so that an 8xA100 GPU cluster can look to the user like a single GPU with 640 GB of memory.
Profiling GPU code
It is more difficult to profile GPU programs since half of the code is running on the CPU and half on the GPU. From outside the program, if a user can ssh into the compute node then running nvidia-smi provides a snapshot of the GPU utilization and GPU memory usage. In the job script the nvprof function can be used in place of the time function to give profile information for various functions of the job.
Compiling and running GPU jobs
Depending on your system you may need to load a module to gain access to the nvcc compiler for CUDA code. nvcc –help can provide you with all the optional parameters, but a basic compile is like nvcc code.cu -o cuda_exec_name. You can then run the executable like any other except that there must be a GPU present. If you are on an HPC system with the Slurm scheduler, you can request a single GPU by adding the –gres=gpu:1 parameter. You can also request a specific type of GPU, but this depends on how each system is set up so you will need to refer to the user documentations. On our HPC cluster at Kansas State University the request for one NVIDIA RTX 3090 would be –gres=gpu:geforce_rtx_3090:1.
Run a simple GPU job if you have access to an HPC system with GPUs
Compile and run the hello_from_gpu.cu CUDA program. You may need to load a module to gain access to the nvcc compiler. There is an sb.hello_from_gpu Slurm batch script to submit the job.
The goal is just to start getting you comfortable with compiling and submitting GPU jobs. Use nvcc hello_gpu.cu -o hello_from_gpu to compile. If you have Slurm on your HPC system you can submit the job with sbatch sb.hello_gpu but you may need to add a partition.
Summary
GPUs are more difficult to program, so for most scientists you will only want to know how to run codes when GPUs are involved and not how to program them yourself. If you are fortunate enough to have an algorithm that someone has accelerated with a GPU, GPUs can often provide an order of magnitude increase in speed over just using CPUs and greatly reduce the cost of doing a calculation.
- Start to become comfortable with using GPUs on an HPC system.
Content from High-Throughput Computing
Last updated on 2024-08-13 | Edit this page
Estimated time: 15 minutes
Overview
Questions
- What is the difference between high-throughput computing and cluster computing?
Objectives
- Learn what types of jobs run well on HTC systems.
- Understand how to submit jobs using OSG.
High-Throughput Computing
A typical HPC system today is a cluster computer made of one or more head nodes and many compute nodes tied together with a common file server and having a batch scheduler to control where jobs run. Some of the largest supercomputers in the world are similar conceptually to a cluster computer but have more custom designed circuit boards as nodes and may be packaged up and racked in a different manner than cluster systems that use more off-the-shelf components.
A High-Throughput Computer (HTC) system is designed to run very large numbers of small jobs and is often made up of many HPC systems separated geographically. In the U.S. the main example is the Open Science Grid (OSG) which is open for use to anyone associated with a U.S. university or national laboratory. Time is given away freely on a first-come first-served basis and computer resources are donated by various institutions. Since HTC is geared toward running large numbers of smaller jobs, the jobs fit well into the spare nooks and crannies of most HPC systems and these systems are set up to kill any OSG job when the host institute needs those resources back. In this way the HTC jobs run invisibly on the over 100 HPC sites spread across the U.S. offering free computing to any who need to run small jobs while greatly enhancing the usage of each HPC system by using the spare CPU cycles that would otherwise go wasted.
Large numbers of small jobs
One of the first examples of HTC was the SETI program where individual PC users could donate their CPU cycles to the project to search through large quantities of data to try to find signals that might come from intelligent life. This problem is ideal for HTC since it can be broken up into large numbers of small jobs, and any job that did not return an answer in a given period was just thrown away and rerun on another system.
OSG has the ability to run a wide variety of jobs including large memory and GPU runs, but it is much more difficult than running the simple small jobs that it was designed for. The OSG guidelines for the typical job are 1-8 cores, up to 40 GB memory and 10 GB IO, and up to 20 hours run-time.
The most important aspect of working with HTC systems is that the jobs be self-contained as much as possible, and be able to run on any operating system and use mainstream modules. This is essential since each job may be run on a wide range of HPC environments. Executables that are dynamically linked can work if you request the matching resources by specifying things like acceptable CPU architectures. Statically linked executables will always work. Containers take more effort to set up but are ideal for HTC since all executables, modules, and environmental variables are set within the container. Running on multiple nodes using MPI is possible but difficult usually requiring containers with the MPI connections specified externally.
Using OSG
Users may submit jobs to the OSG queue through their institute’s portal if one exists or through the OSG Connect submission service at the link below. If using the OSG Connect portal you will need to request access and arrange for a short Zoom meeting with one of their support staff. There are links to the OSG Consortium and support documentation at the end of this lesson.
Working with the HTCondor scheduler
Submitting jobs to HTCondor for scheduling on any of the hundreds of remote systems available is similar to using any scheduler, except for the notes above on the job not relying on modules and libraries that may not be available or labeled the same everywhere. Below is an example job script to run the stand-alone executable namd2. Note that the X64_64 CPU architecture is specified. The script requests 1 GB of disk for this job and directs that all files from the directory input_files be transferred along with the job at the start, and all files in output be transferred back at the end. The queue 1 command then specifies the number of these jobs to submit. If multiple jobs are submitted then the environmental variable $(Process) will be used in the script to differentiate each with that being between zero and the number of jobs specified minus one.
BASH
#!/bin/bash -l
output = osg.namd.out
error = osg.namd.error
log = osg.namd.log
# Requested resources
request_cpus = 8
request_memory = 8 GB
request_disk = 1 GB
requirements = Arch == "X86_64" && HAS_MODULES == True
transfer_input_files = input_files/ # Slash means all files in that directory
executable = namd2
arguments = +p8 test.0.namd
transfer_output_files = output
queue 1Most systems will have support for modules, but the HAS_MODULES == True requirement can mean some systems are not supported. Most systems use RHEL7, so specifying that may also rule out use of RHEL6 and RHEL8 systems. In general, if you use a mainstream operating system and modules then you should be fine. Otherwise you likely will need to use a container.
Once you have the script you can submit, monitor, and control the job using commands like those below.
BASH
> condor_submit htc_job.sh # Submit the condor script to the queue
> condor_q # Check on the status while in the queue
> condor_q netid # Check status of currently running jobs
> condor_q 121763 # Check status of a particular job
> condor_history 121763 # Check status of a job that is completed
> condor_history -long 121763 # Same but report more info
> condor_rm 121763 # Remove the job number specified
> condor_rm daveturner # Remove all jobs for the given usernameHTC Outside the U.S.
Need to add info about non-US HTC systems
Optional Homework: Get an OSG account and submit a test job.
If you are in the U.S. and want a big challenge, request an OSG account and try submitting some small jobs.
Summary
High-Throughput Computing is free in the U.S. using the Open Science Grid. It is great for running large numbers of small jobs. Using it for GPU jobs or when large memory or IO is needed is possible but much more challenging. Aside from that it is similar to running jobs using any other scheduler.
- Explore the basics of HTC computing.
Content from HPC Resources
Last updated on 2024-08-13 | Edit this page
Estimated time: 15 minutes
Overview
Questions
- How do I get access to HPC resources?
Objectives
- Understand what HPC resources are available in the U.S.
- Learn how to access these resources for free.
Getting Access to HPC Resources
It is very typical to start a project on a personal computer only to find that you need more performance either as computing power or memory. The next step may be to run your code on a more powerful workstation such as a departmental server. But when you still need more performance where do you go? Most scientists do not understand that there are many options for getting more powerful computing resources, and that they are commonly free for the asking.
If you are in a university or laboratory environment the first place to look is your local supercomputing center if you have one. While many provide priority access to the compute resources to those that provide research funds, most also have some means of providing significant resources to users that can’t provide financial support. Many university cluster computers work on the condo model where scientific groups can purchase compute nodes that they have full priority on, but in return for the university managing those compute nodes anyone else is allowed to use them when idle. This pay for priority model works well in sharing computing resources between the haves and the have nots.
In many states one of the larger universities may provide free access to scientists in smaller universities in the state. Kansas State University for example provides free access to anyone associated with any university in the state. Many other states provide funding for a single supercomputing center that all in the state can use.
Remote resources
There are many national supercomputing centers in the U.S. that receive federal funding from the National Science Foundation (NSF) and Department of Energy (DoE). These supercomputers can cost $10-$100 million or above and have capacities now over 1 exa-flops (billion billion floating-point operations per second). When these are funded, part of the compute cycles are designated for access to scientists not directly associated with those institutions.
Account and allocation requests as well as access is handled through user portals. Most of these remote systems have startup accounts available that may typically be in the range of 5,000 core-hours (5000 hours on 1 core or 500 hours on 10 cores for example). Getting access is as easy as knowing where and how to ask.
The ACCESS portal to remote NSF supercomputers
The ACCESS portal is formerly known as XSEDE. The link below is where to go to request an account and allocation, submit a proposal, and find documentation and support. There are a very wide variety of supercomputing systems available to users associated with a university or laboratory. Small startup allocations usually only take a few minutes to apply for and are typically approved within a few days.
Many universities have a Campus Champion with a larger allocation on many systems designed to be shared across a campus. When these allocations run out, they can simply request more but they are intended to get users needing long-term support the experience on a system so that they can submit a full proposal. While startup requests are typically on a specific system and may be around 5,000 core-hours, campus champions may be granted ~5 times as much plus access to GPU and large memory nodes. If you are on a campus with a Campus Champion then you just need to apply for an account then contact them to get access to the actual allocations.
Follow the ACCESS link at the end of this lesson for a complete list of supercomputing resources, but below is an example of some of the systems available (last updated August of 2022).
Bridges2 at the Pittsburgh Supercomputing Center (PSC)
488 RM Regular Memory compute nodes
- 2 x AMD EPYC 7742 –> 128 cores
- 256 GB memory (16 more nodes have 512 GB each)
- 3.84 TB NVMe SSD
- Mellanox HDR 200 Gbps network
4 EM Extreme Memory compute nodes
- 4 x Intel Cascade 8260M –> 96 cores
- 4 TB memory
- 7.68 TB NVMe SSD
- Mellanox HDR 200 Gbps network
24 GPU compute nodes
- 2 x Intel Gold Cascade 6248 –> 40 cores
- 8 x NVIDIA Tesla v100 32 GB sxm2 GPU cards
- 512 GB memory
- 7.68 TB NVMe SSD
- Mellanox HDR 200 Gbps network
Expanse at the San Diego Supercomputing Center (SDSC)
728 compute nodes
- 2 x AMD EPYC 7742 –> 128 cores
- 256 GB memory
4 Large Memory nodes
- 4 x Intel Cascade 8260M –> 96 cores
- 2 TB memory
52 GPU nodes
- 2 x Intel Xeon 6248 –> 40 cores
- 4 x NVIDIA Tesla v100 GPU cards
- 384 GB memory
Cluster-wide capabilities
- 12 PetaByte Lustre file system
- 7 PetaByte CEPH object store
- 56 Gbps bi-directional HDR InfiniBand network
National Energy Research Scientific Computing Center (NERSC)
While allocations for the NSF supercomputing centers are managed through the ACCESS website, external access to the Department of Energy (DoE) supercomputers are managed through NERSC with the website link at the end of this module. Many DoE supercomputers can be accessed by scientists in universities and laboratories, but the process is more involved. Access is still free, but you typically need to write a proposal for significant node-hours and fully justify that the science you intend to do is important and fits within the DoE mission, and demonstrate that your code will use the requested resources efficiently.
Open Science Grid (OSG)
If you need to run many smaller jobs, then High-Throughput Computing (HTC) covered in the previous chapter may be ideal for you. OSG access is fairly easy for users in the U.S. and provides virtually unlimited access.
National Research Platform (NRP)
The NRP is a partnership of over 50 institutions led by UC San Diego supported in large part by NSF. The Nautilus system is a HyperCluster for running containerized big data applications using kubernetes for the container management. This is a distributed set of compute nodes similar to OSG but applications must specifically be self-contained and are guaranteed isolated access to the resources allocated.
While kubernetes is more difficult for the average user, this system provides access to much more powerful computing including very high-end GPUs. The compute nodes are mainly GPU-based and research is aimed at Machine Learning codes.
Cloud Computing
Running HPC jobs in the cloud is much more difficult and costly than many people understand. Most cloud computing vendors do give small amounts of access away for free on a trial basis for those that want to experiment. The amount is very minimal in the context of running an HPC job.
As of August of 2024, Google Cloud Platform (GCP) offered $300 credit to new customers for example. Amazon Web Services (AWS) offered several free trial plans with $750 credits. Microsoft Azure offers a $200 credit for 30 days for eligible new customers.
Summary
Many projects that start on a laptop or desktop system end up needing larger resources. Transitioning to an HPC system can be challenging, but the best news is that HPC resources are often free for the asking. You just need to know where and how to ask, and hopefully this module will give you ideas on where to start.
- There are many HPC resources that are totally free for the asking.